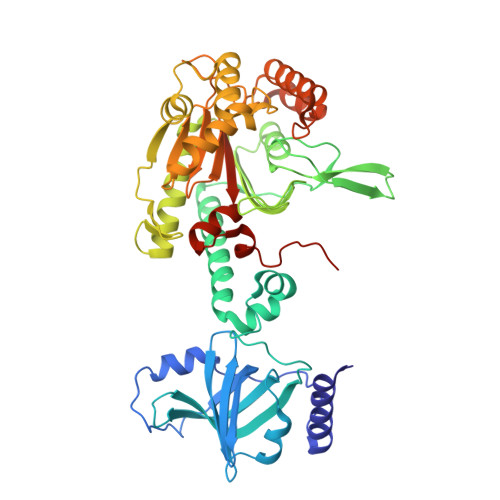
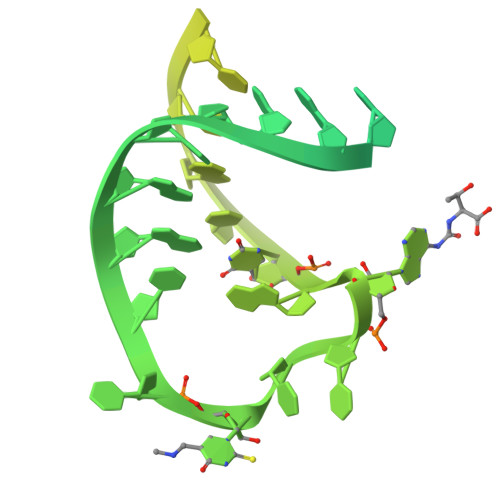
The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNA(Lys) and a T. thermophilus tRNA(Lys) transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue.
Cusack, S., Yaremchuk, A., Tukalo, M.(1996) EMBO J 15: 6321-6334
- PubMed: 8947055
- Primary Citation of Related Structures:
9SLZ - PubMed Abstract:
The crystal structures of Thermus thermophilus lysyl-tRNA synthetase, a class IIb aminoacyl-tRNA synthetase, complexed with Escherchia coli tRNA(Lys)(mnm5 s2UUU) at 2.75 A resolution and with a T. thermophilus tRNA(Lys)(CUU) transcript at 2.9 A resolution are described. In both complexes only the tRNA anticodon stem-loop is well ordered. The mode of binding of the anticodon stem-loop to the N-terminal beta-barrel domain is similar to that previously found for the homologous class IIb aspartyl-tRNA synthetase-tRNA(Asp) complex except in the region of the wobble base 34 where either mnm5 s2U or C can be accommodated. The specific recognition of the other anticodon bases, U-35 and U-36, which are both major identity elements in the lysine system, is also described. Additional crystallographic data on a ternary complex with a lysyl-adenylate analogue show that binding of the intermediate induces significant conformational changes in the vicinity of the active site of the enzyme.
- European Molecular Biology Laboratory, Grenoble Outstation, France.
Organizational Affiliation: