Structural remodeling of the mitochondrial protein biogenesis machinery upon proteostatic stress
Ehses, K., Lopez-Alonso, J.P., Gonzalez, A., Tascon, I., Ubarretxena-Belandia, I., Fernandez-Busnadiego, R.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60 kDa heat shock protein, mitochondrial | 527 | Homo sapiens | Mutation(s): 0 Gene Names: HSPD1, HSP60 EC: 5.6.1.7 | 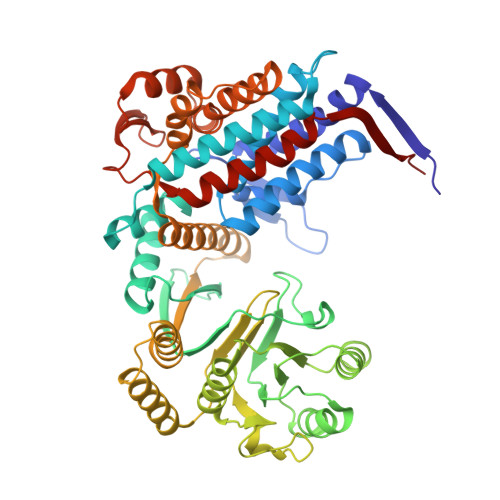 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P10809 (Homo sapiens) Explore P10809 Go to UniProtKB: P10809 | |||||
PHAROS: P10809 GTEx: ENSG00000144381 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P10809 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP (Subject of Investigation/LOI) Query on ATP | H [auth A] K [auth B] N [auth C] Q [auth D] T [auth E] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| K (Subject of Investigation/LOI) Query on K | BA [auth G] J [auth A] M [auth B] P [auth C] S [auth D] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | AA [auth G] I [auth A] L [auth B] O [auth C] R [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 4.1.2 |
| MODEL REFINEMENT | PHENIX | 1.21rc1_4933 |
| MODEL REFINEMENT | Coot | 0.9.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministerio de Ciencia e Innovacion (MCIN) | Spain | PID2022-143177NB-I00 |