Identification of an allosteric site on the E3 ligase adapter cereblon
Chung, C.(2026) Nature
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA damage-binding protein 1 | 1,148 | Homo sapiens | Mutation(s): 0 Gene Names: DDB1, XAP1 | 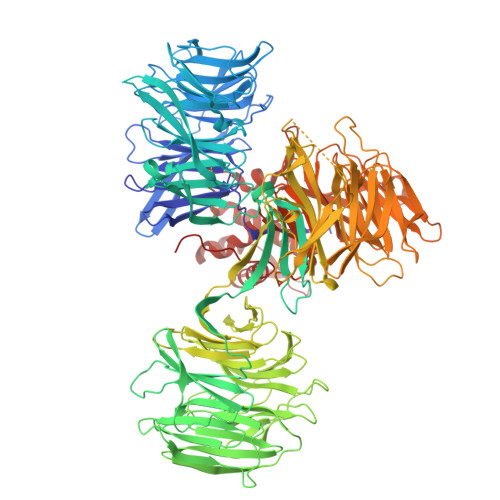 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q16531 (Homo sapiens) Explore Q16531 Go to UniProtKB: Q16531 | |||||
PHAROS: Q16531 GTEx: ENSG00000167986 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q16531 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein cereblon | 382 | Homo sapiens | Mutation(s): 0 Gene Names: CRBN, AD-006 | 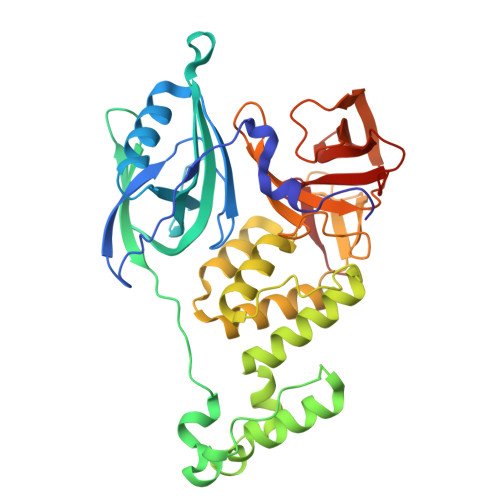 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96SW2 (Homo sapiens) Explore Q96SW2 Go to UniProtKB: Q96SW2 | |||||
PHAROS: Q96SW2 GTEx: ENSG00000113851 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96SW2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1CEG (Subject of Investigation/LOI) Query on A1CEG | K [auth B] | N-[3-(benzyloxy)pyridin-2-yl]-N'-(4-cyano-2-hydroxyphenyl)urea C20 H16 N4 O3 NFPDKHFRABMOJP-UHFFFAOYSA-N |  | ||
| LVY Query on LVY | J [auth B] | S-Lenalidomide C13 H13 N3 O3 GOTYRUGSSMKFNF-JTQLQIEISA-N |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | H [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | C [auth A] D [auth A] E [auth A] F [auth A] G [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 72.16 | α = 90 |
| b = 129.518 | β = 90 |
| c = 198.797 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |