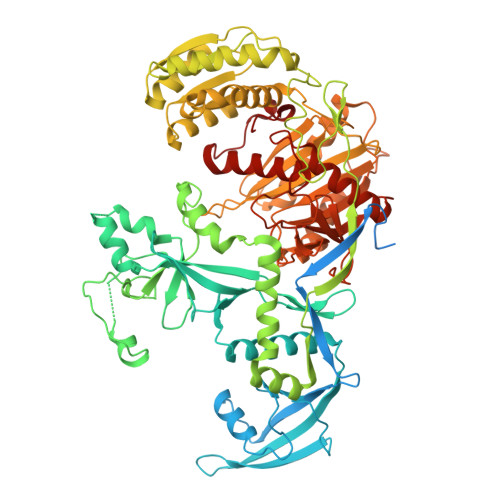
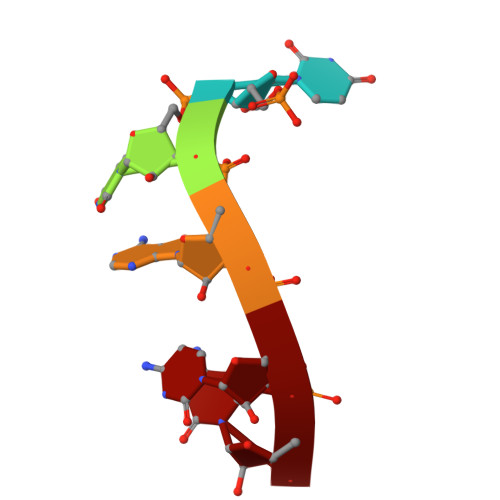
Structure of the MIWI endoribonuclease bound to pachytene piRNAs from mouse testes.
Raad, N., Fernandez-Rodriguez, C., Pandey, R.R., Mohammed, I., Uchikawa, E., Burger, F., Homolka, D., Pillai, R.S.(2026) Cell Rep 45: 116804-116804
- PubMed: 41528842
- DOI: https://doi.org/10.1016/j.celrep.2025.116804
- Primary Citation of Related Structures:
9S0Z, 9S1E, 9SHP, 9SHQ - PubMed Abstract:
PIWI-interacting RNAs (piRNAs) guide PIWI endoribonucleases to destroy transposon transcripts, ensuring animal fertility. Here, we report the cryo-electron microscopy structure of the MIWI-pachytene piRNA complex isolated from mouse testes. The piRNA is held via non-specific charge-based interactions with the RNA backbone and by specific recognition of the first nucleotide uridine by residues within the MID and PIWI domains. The first six nucleotides of the guide RNA take up the A-form conformation to facilitate pairing with the target. The RNA channel is wider than that observed in insect PIWI proteins, explaining the tolerance for piRNA seed:target mismatches. The PIWI endonuclease domain is in an inactive "un-plugged" state, with the loop containing a catalytic residue (E671) requiring structural re-orientation for activity. Furthermore, the PIWI domain reveals a conserved pre-formed pocket that may serve to accommodate a conserved tryptophan from the interacting factor GTSF1 to promote small RNA-guided endoribonuclease activity.
- Department of Molecular Biology, Science III, University of Geneva, 30 Quai Ernest-Ansermet, Geneva 1211, Switzerland. Electronic address: nicole.raad@unige.ch.
Organizational Affiliation: