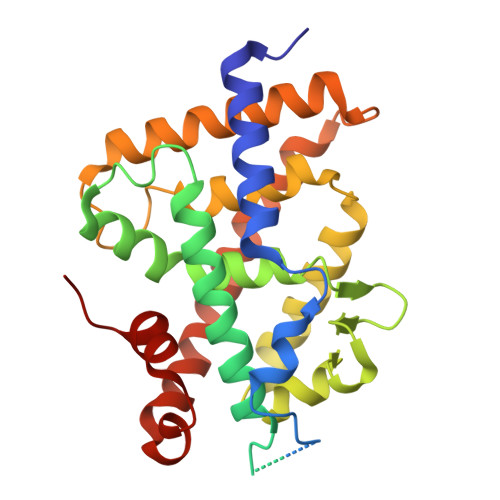
Hyperagonism of a Vitamin D Receptor Agonist/Histone Deacetylase Inhibitor Hybrid Molecule.
Sarmadi, F., Caza, A., Gao, Z., Rochel, N., Gleason, J.L., White, J.H.(2025) J Med Chem 68: 20675-20688
- PubMed: 40968099
- DOI: https://doi.org/10.1021/acs.jmedchem.5c01932
- Primary Citation of Related Structures:
9RCG - PubMed Abstract:
1,25-Dihydroxyvitamin D (1,25D) analogs engage the vitamin D receptor (VDR) and can exert anticancer and immunomodulatory effects. Although tumors often resist 1,25D monotherapy, combining VDR agonism with histone deacetylase inhibitors (HDACi) restores anticancer efficacy. Here, we present AC-340, a novel bifunctional molecule that incorporates HDACi into a VDR agonist backbone. Besides its robust bifunctionality in vitro in multiple melanoma models, RNaseq analysis of B16-F10 mouse melanoma cells revealed that AC-340 superinduces the expression of a broad array of VDR target genes. Comparative structural studies and ChIP-qPCR revealed that AC-340 forms more interactions than 1,25D with residues in the VDR coactivator binding domain, leading to more efficacious recruitment of coactivator CBP. This, likely coupled with AC-340 HDACi activity, leads to elevated H3K27 acetylation of VDR target genes, a mark of active transcription. Thus, AC-340 functions as a VDR hyperagonist and should be efficacious in mono- or combination therapies against multiple cancer models.
- Department of Physiology, McGill University, 3655 Promenade Sir William Osler, Montreal, QC H3G 1Y6, Canada.
Organizational Affiliation: