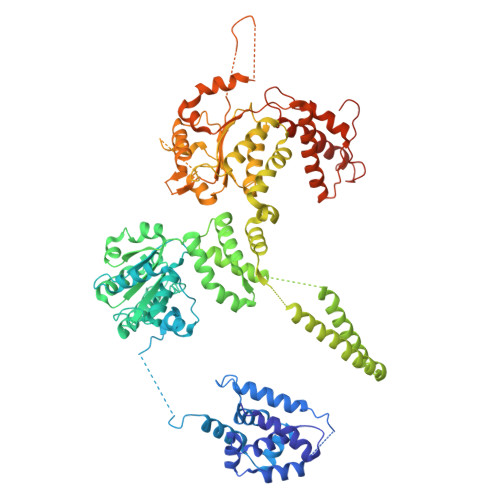
Structure of the central Staphylococcus aureus AAA+ protease MecA/ClpC/ClpP.
Azinas, S., Wallden, K., Katikaridis, P., Jenne, T., Schahl, A., Mogk, A., Carroni, M.(2025) Commun Biol 8: 1467-1467
- PubMed: 41087538
- DOI: https://doi.org/10.1038/s42003-025-08908-w
- Primary Citation of Related Structures:
9R2S, 9RAI - PubMed Abstract:
Bacterial AAA+ proteases are composed of a AAA+ partner (e.g., ClpC) and an associated peptidase (e.g., ClpP). They represent ATP-fuelled and self-compartmentalized proteolytic machines that are crucial for stress resistance and virulence. ClpC requires cooperation with adaptor proteins such as MecA for activation and complex formation with ClpP. Here, we present the cryo-EM structure of the MecA/ClpC/ClpP complex from the major pathogen Staphylococcus aureus. MecA forms a dynamic crown on top of the ClpC/ClpP complex with its substrate-binding domain positioned near the ClpC pore site, likely facilitating substrate transfer. ClpC/ClpP complex formation involves ClpC P-loops and ClpP N-terminal β-hairpins, which insert into the central ClpC threading channel and contact sites next to the ClpC ATPase center. ClpC and ClpP interactions are asymmetric and dictated by the activity states of ClpC ATPase subunits. ClpP binding increases ClpC ATPase and threading activities in a β-hairpin dependent manner, illuminating an allosteric pathway in the cooperation of ATPase and peptidase components in bacterial AAA+ proteases.
- Science for Life Laboratory, Department of Biochemistry and Biophysics, Stockholm University, Stockholm, Sweden.
Organizational Affiliation: