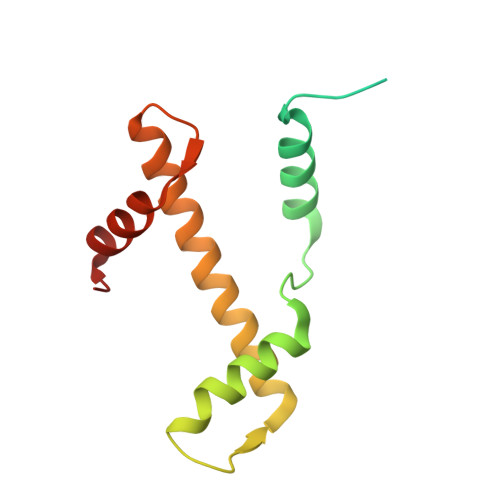
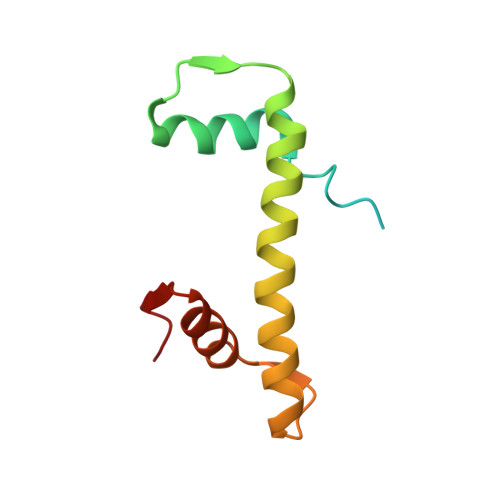
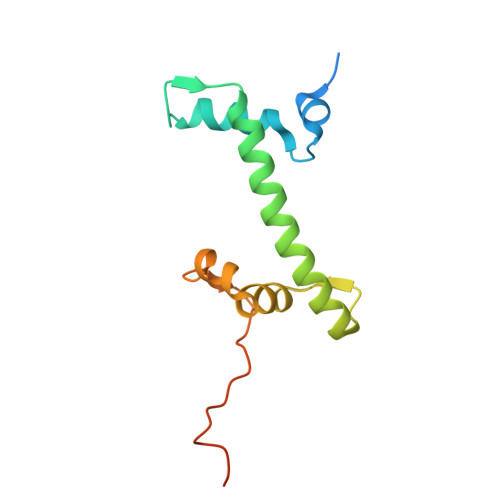
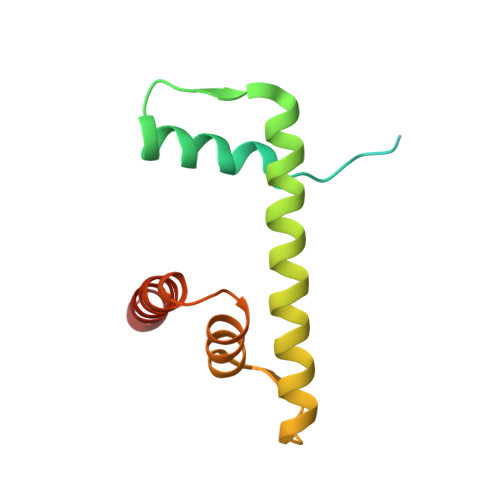
Structural characterisation of chromatin remodelling intermediates supports linker DNA-dependent product inhibition as a mechanism for nucleosome spacing.
Hughes, A.L., Sundaramoorthy, R., Owen-Hughes, T.(2025) Elife 14
- PubMed: 41439750
- DOI: https://doi.org/10.7554/eLife.52513
- Primary Citation of Related Structures:
9R5K, 9R5S, 9R5W - PubMed Abstract:
Previously, we showed that Saccharomyces cerevisiae Chd1 chromatin remodelling enzyme associates with nucleosomes oriented towards the longer linker (Sundaramoorthy et al., 2018) (1). Here, we report a series of structures of Chd1 bound to nucleosomes during ongoing ATP-dependent repositioning. Combining these with biochemical experiments and existing literature, we propose a model in which Chd1 first associates oriented to sample putative entry DNA. In an ATP-dependent reaction, the enzyme then redistributes to the opposite side of the nucleosome, where it subsequently adopts a conformation productive for DNA translocation. Once this active complex extends the nascent exit linker to approximately 15 bp, it is sensed by the Chd1 DNA binding domain, resulting in conversion to a product-inhibited state. These observations provide a mechanistic basis for the action of a molecular ruler element in nucleosome spacing.
- Molecular Cell and Developmental Biology, School of Life Sciences, University of Dundee, Dundee, United Kingdom.
Organizational Affiliation: