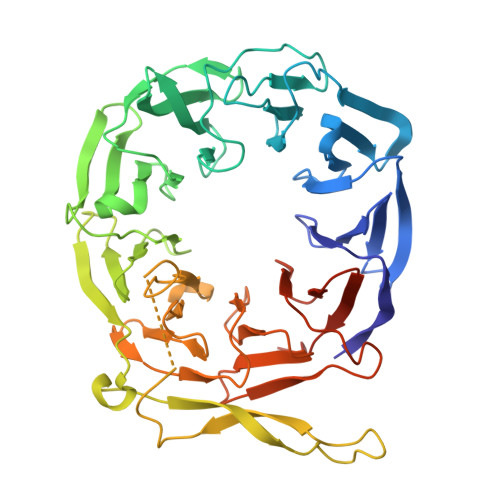
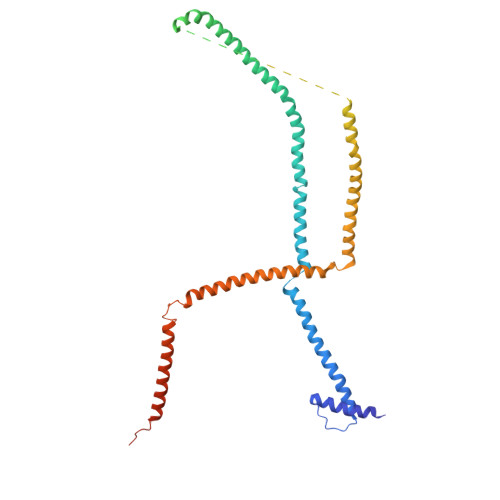
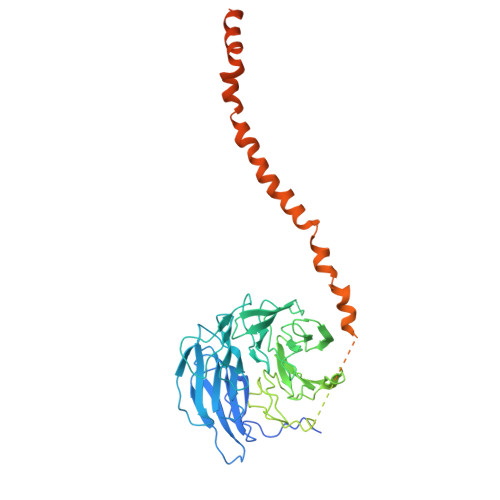
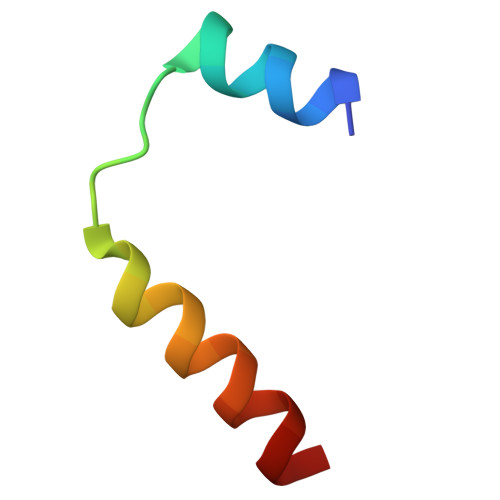
Structure and assembly of the A-C linker connecting microtubule triplets in centrioles.
Cai, B., Xu, J., Collet, E.H., Aarts, E., Luo, L., Leitner, A., Ishikawa, T., Beltrao, P., Pearson, C.G., Pilhofer, M., Wieczorek, M.(2025) Sci Adv 11: eady3689-eady3689
- PubMed: 41061066
- DOI: https://doi.org/10.1126/sciadv.ady3689
- Primary Citation of Related Structures:
9QZC, 9QZF - PubMed Abstract:
Centriole assembly involves the coordination of centriolar modules. One module is the A-C linker, an enigmatic protein assembly connecting the A-microtubule of one microtubule triplet to the C-microtubule of the neighboring triplet. Here, we integrated biochemistry, multiscale cryo-electron microscopy, and AlphaFold modeling to investigate the architecture of the centriole. Using an improved centriole isolation method, we determined the structure of the A-C linker bound to microtubule triplets, which revealed how the A-C linker cross-links microtubules and integrates with the B-C junction. We found marked changes in the structure and composition of the A-C linker that correlate with the presence of other centriolar modules, including the pinhead, cartwheel, and inner scaffold. Our findings show that the A-C linker is a highly integrated component of the centriole whose polymorphism may orchestrate the assembly of spatially distinct centriolar modules, and provide a framework for dissecting the biology of centrioles.
- Institute of Molecular Biology and Biophysics, Department of Biology, ETH Zürich, Zürich, Switzerland.
Organizational Affiliation: