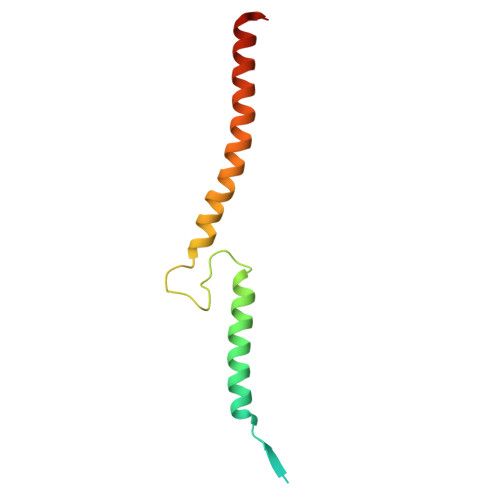
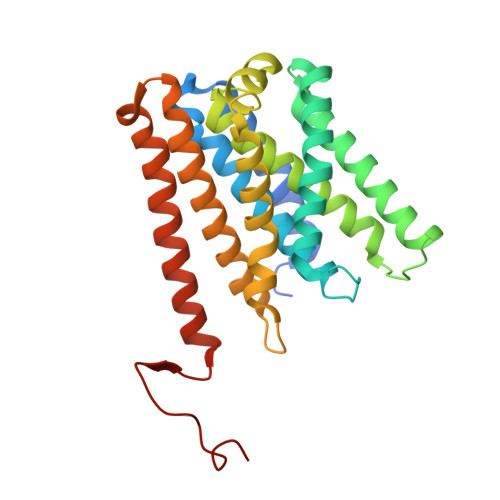
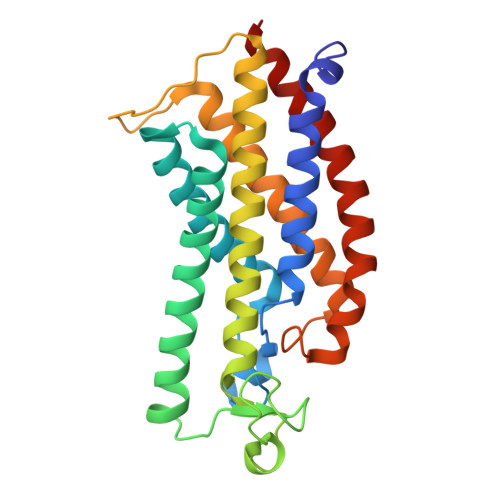
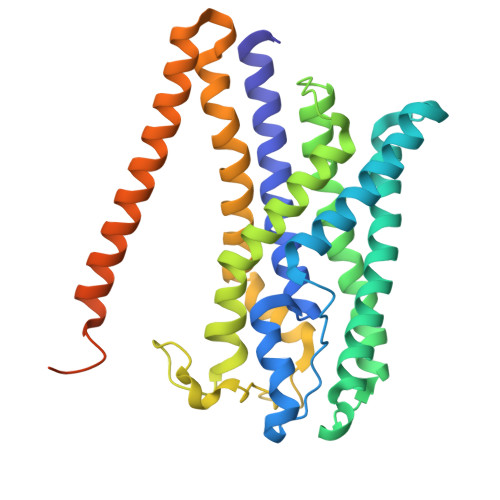
Structure of the Methanosarcina mazei Mtr complex bound to the oxygen-stress responsive small protein MtrI.
Reif-Trauttmansdorff, T., Herdering, E., Bohn, S., Pascoa, T., Kahnt, J., Zimmer, E., Kumar, A., Schmitz, R.A., Schuller, J.M.(2025) Nat Commun 17: 133-133
- PubMed: 41436738
- DOI: https://doi.org/10.1038/s41467-025-67705-5
- Primary Citation of Related Structures:
9QTP, 9QTQ, 9QTR, 9QTS - PubMed Abstract:
Methanogenic archaea emit ~1 Gt of methane annually, impacting global carbon cycling and climate. Central to their energy metabolism is a membrane-bound, sodium-translocating methyltransferase complex: the N⁵-tetrahydromethanopterin:CoM-S-methyltransferase (Mtr). It couples methyl transfer between two methanogen-specific cofactors with sodium ion transport across the membrane, forming the only energy-conserving step in hydrogenotrophic methanogenesis. Here, we present a 2.1 Å single-particle cryo-EM structure of the Mtr complex from Methanosarcina mazei. The structure reveals the organization of all catalytic subunits, embedded archaeal lipids and the sodium-binding site. Most strikingly, we discover MtrI, a previously unannotated small open-reading frame encoded protein ( < 100 aa) found within the order of Methanosarcinales that binds both the top of the sodium-channel and cytosolic domain of MtrA via its cobamide cofactor in response to oxygen exposure. This interaction likely prevents sodium leakage and stabilizes the complex under oxidative conditions, revealing an unexpected regulatory mechanism in methanogen energy conservation.
- Center for Synthetic Microbiology (SYNMIKRO) Research Center and Department of Chemistry, Philipps-Universität Marburg, Karl-von-Frisch Straße 14, Marburg, Germany.
Organizational Affiliation: