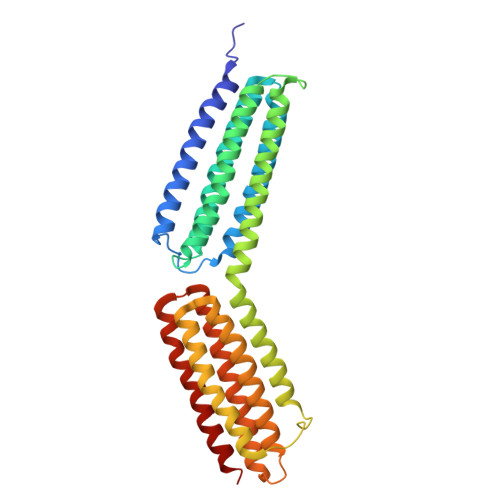
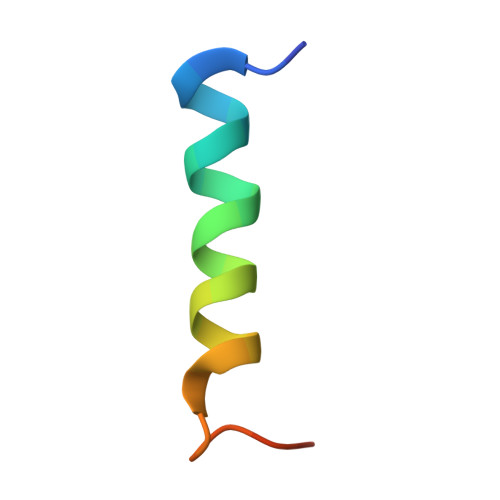
Talin-tensin3 interactions regulate fibrillar adhesion formation and tensin3 phase separation.
Li, X., Konstantinou, R., Meena, V.K., Notash, S., Khalil, K., Whalley, T., Atherton, P., Barsukov, I., Zacharchenko, T., Ballestrem, C.(2026) J Cell Biol 225
- PubMed: 41269158
- DOI: https://doi.org/10.1083/jcb.202503155
- Primary Citation of Related Structures:
9QN7 - PubMed Abstract:
Integrin-mediated cell-matrix adhesions regulate communication between cells and the extracellular matrix. In matrix-secreting cells, fibrillar adhesions (FBs) containing high levels of α5β1 integrins and the tensin3 adaptor protein are essential for fibronectin (FN) fibrillogenesis. Here, we demonstrate that tensin3 binds to four helical regions (R3, R4, R8, and R11) of talin, the principal integrin activator. Structural analysis revealed the residues critical for the tensin3-talin interaction, and mutational analysis showed that talin R8 and R11 are essential for FB formation and FN fibrillogenesis. Cellular experiments demonstrate that tensin3 binding to talin not only regulates integrin activation, but also modulates tensin3's propensity to undergo liquid-liquid phase separation (LLPS). Formation of such LLPS condensates increased when cells were plated on soft substrates compared with stiff ones. This effect was abolished by blocking the interaction between tensin3 and talin. Our data suggest a model in which LLPS condensates provide a signaling platform involved in cellular responses to sudden changes in tissue mechanics.
- Faculty of Biology, Medicine and Health, Wellcome Centre for Cell-Matrix Research, University of Manchester , Manchester, UK.
Organizational Affiliation: