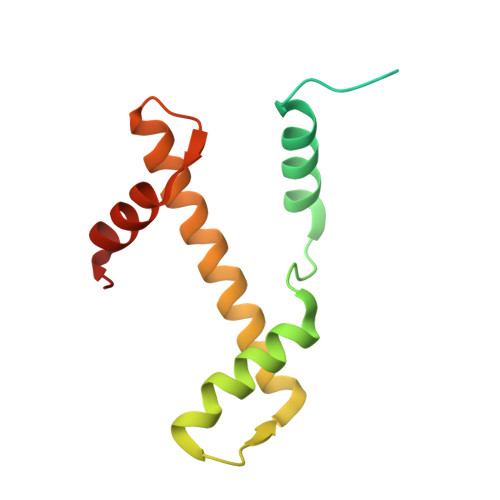
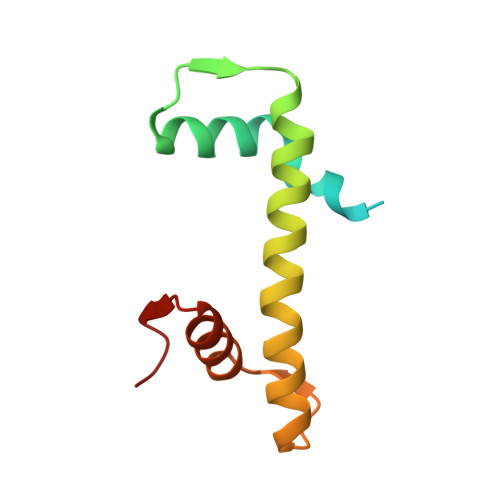
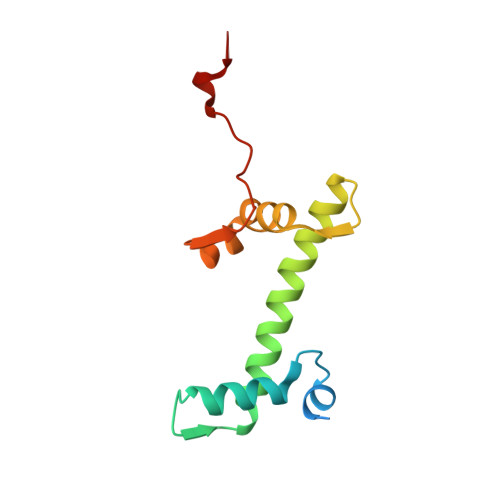
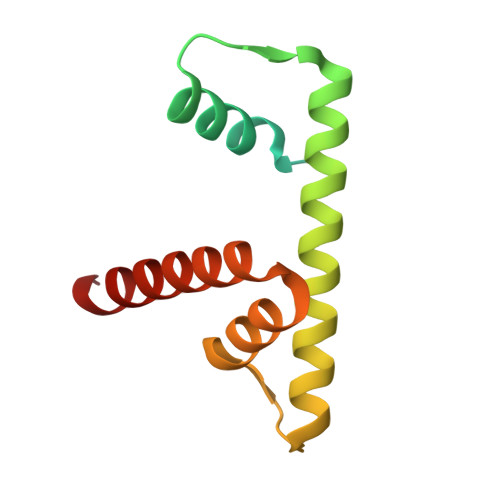
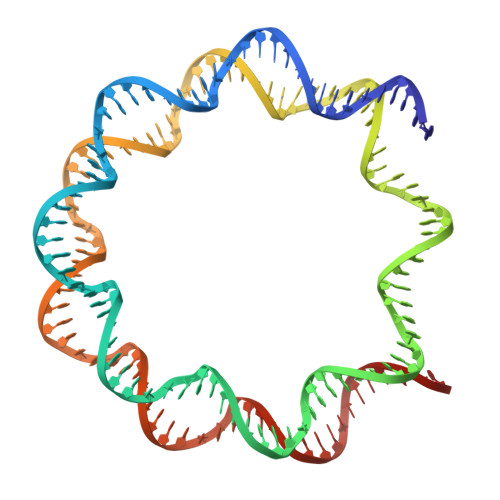
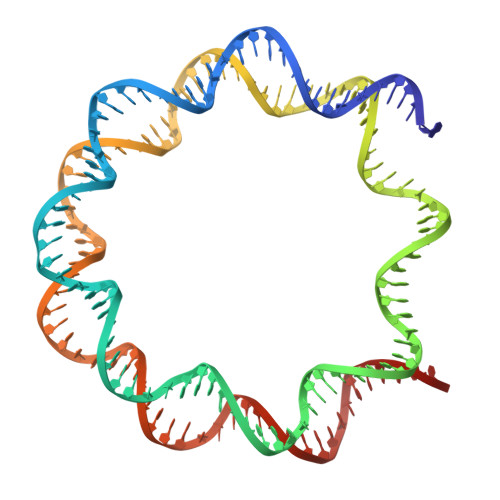
Structure of the nucleosome-bound human BCL7A.
Martin, F., Kazrani, A.A., Lafouge, J., Diaz-Jimenez, D.M., Siebert, S., Fabbro-Burtschell, L., Maillard, E., Lapouge, K., Mertens, H.D.T., Sauter, C., Leitner, A., Ochsenbein, F., Blais, A., Bergamin, E.(2025) Nucleic Acids Res 53
- PubMed: 40207634
- DOI: https://doi.org/10.1093/nar/gkaf273
- Primary Citation of Related Structures:
9QAJ - PubMed Abstract:
Proteins of the BCL7 family (BCL7A, BCL7B, and BCL7C) are among the most recently identified subunits of the mammalian SWI/SNF chromatin remodeler complex and are absent from the unicellular version of this complex. Their function in the complex is unknown, and very limited structural information is available, despite the fact that they are mutated in several cancer types, most notably blood malignancies and hence medically relevant. Here, using cryo-electron microscopy in combination with biophysical and biochemical approaches, we show that BCL7A forms a stable, high-affinity complex with the nucleosome core particle (NCP) through binding of BCL7A with the acidic patch of the nucleosome via an arginine anchor motif. This interaction is impaired by BCL7A mutations found in cancer. Further, we determined that BCL7A contributes to the remodeling activity of the mSWI/SNF complex and we examined its function at the genomic level. Our findings reveal how BCL7 proteins interact with the NCP and help rationalize the impact of cancer-associated mutations. By providing structural information on the positioning of BCL7 on the NCP, our results broaden the understanding of the mechanism by which SWI/SNF recognizes the chromatin fiber.
- Department of Functional Genomics and Cancer & Department of Integrated Structural Biology, Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), 67400 Illkirch-Graffenstaden, France.
Organizational Affiliation: