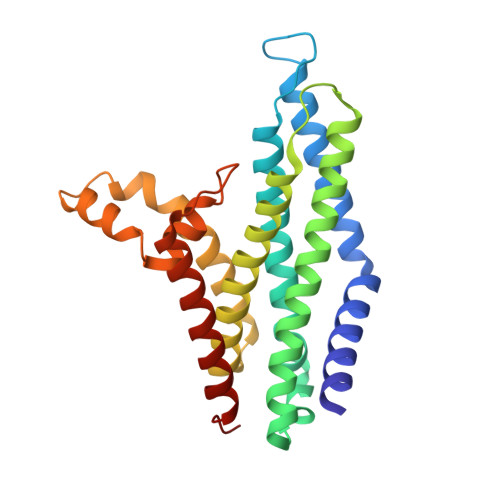
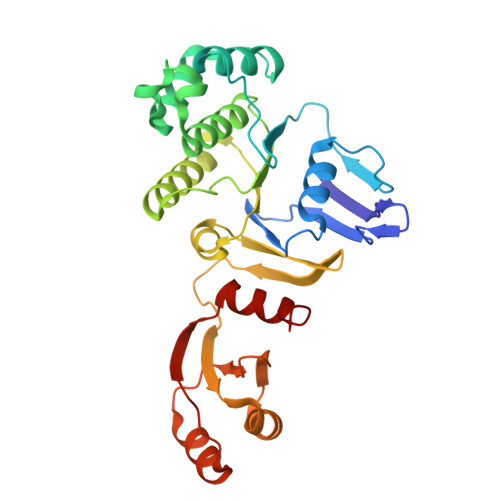
Cryo-EM analysis of the Staphylococcus aureus phenol-soluble modulin exporter PmtCD apo form in detergent micelles, nanodiscs and peptidiscs.
Hu, J., Lazarski, A.C., Li, F.K.K., Worrall, L.J., Burgin, D.J., Zeytuni, N., Dickey, S.W., Otto, M., Strynadka, N.C.J.(2025) Commun Biol 8: 1576-1576
- PubMed: 41249510
- DOI: https://doi.org/10.1038/s42003-025-08955-3
- Primary Citation of Related Structures:
9Q1Y, 9Q2N, 9Q2Q, 9Q2R - PubMed Abstract:
Staphylococci secrete amphipathic peptides known as phenol soluble modulins (PSMs) that play a variety of pathogenic roles including host cell membrane destruction, biofilm development, and the triggering of inflammatory responses. PSM export is facilitated by the essential ATP-binding cassette (ABC) transporter PmtCD, which also provides producer immunity toward the membrane-damaging PSMs. Here, we report cryo-EM structures of PmtCD in a nucleotide-free state using different membrane mimetics - detergent, nanodisc and peptidisc - all featuring the transmembrane domains in an open state with a remarkably expansive intervening lumen. The consistently sized lumen suggests the possibility for two α-helical amphipathic PSMs to pack and passage within. A continuous hydrophobic surface with no apparent single high affinity site is in keeping with the ability of PmtCD to export a variety of hydrophobic PSM peptide substrates. The ATP driven collapse of the PmtD lumen is consistent with the lateral access and extrusion mechanisms of related ABC transporters that translocate membrane embedded substrates. Along with a new ADP product complex and prior ATPγS-bound form, these structures provide insights into the export of PSMs and a foundation for design of trojan horse antimicrobials that target MRSA strains from within by blocking membranolytic PSM export.
- Department of Biochemistry and Molecular Biology, The University of British Columbia, Vancouver, BC, Canada.
Organizational Affiliation: