Yeast Display Reveals Plentiful Mutations That Improve Fusion Peptide Vaccine-Elicited Antibodies Beyond 59% HIV-1 Neutralization Breadth.
Franca, C.T., Pletnev, S., Madan, B., Katsamba, P.S., McKee, K., Morano, N.C., Zhang, B., Bahna, F., Bylund, T., Lin, B.C., Louder, M.K., Mannepalli, S., Nimrania, R., O'Dell, S., Doria-Rose, N.A., Kwong, P.D., Shapiro, L., Sheng, Z., Zhou, T., DeKosky, B.J.(2025) Vaccines (Basel) 13
- PubMed: 41295471
- DOI: https://doi.org/10.3390/vaccines13111098
- Primary Citation of Related Structures:
9Q0W - PubMed Abstract:
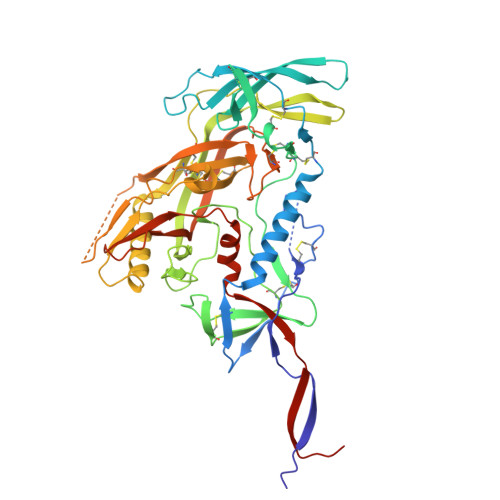
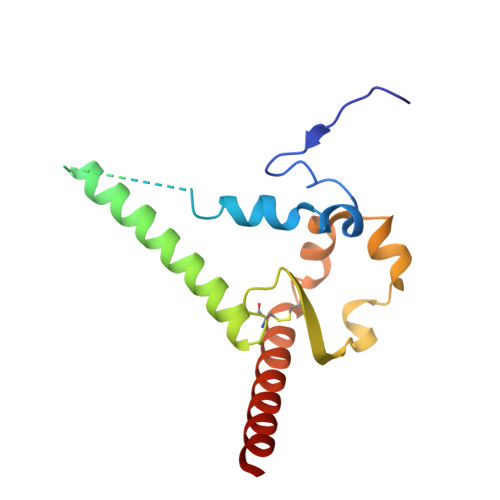
Background/Objectives : Vaccine elicitation of antibodies with high HIV-1 neutralization breadth is a long-standing goal. Recently, the induction of such antibodies has been achieved at the fusion peptide site of vulnerability. Questions remain, however, as to how much anti-fusion peptide antibodies can be improved and whether their neutralization breadth and potency are sufficient to prevent HIV-1 infection. Methods : Here, we use yeast display coupled with deep mutational screening and biochemical and structural analyses to study the improvement of the best fusion peptide-directed, vaccine-elicited antibody, DFPH_a.01, with an initial 59% breadth. Results : Yeast display identified both single and double mutations that improved recognition of HIV-1 envelope trimers. We characterized two paratope-distal light chain (LC) mutations, S10R and S59P, which together increased breadth to 63%. Biochemical analysis demonstrated DFPH-a.01_10R59P-LC, and its component mutations, to have increased affinity and stability. Cryo-EM structural analysis revealed elbow-angle influencing by S10R-LC and isosteric positioning by S59P-LC as explanations for enhanced breadth, affinity, and stability. Conclusions : These results, along with another antibody with enhanced performance (DFPH-a.01_1G10A56K-LC with 64% breadth), suggest that mutations improving DFPH_a.01 are plentiful, an important vaccine insight.
- The Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard University, Cambridge, MA 02139, USA.
Organizational Affiliation: