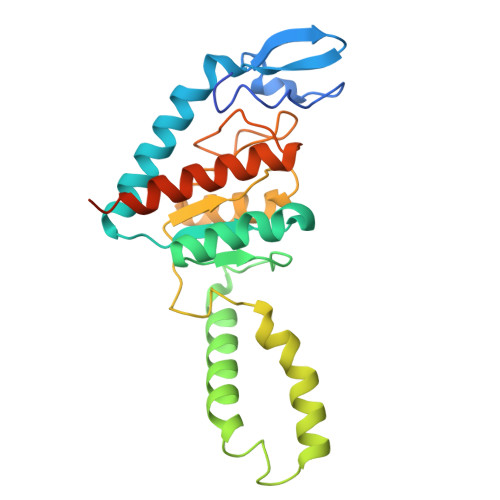
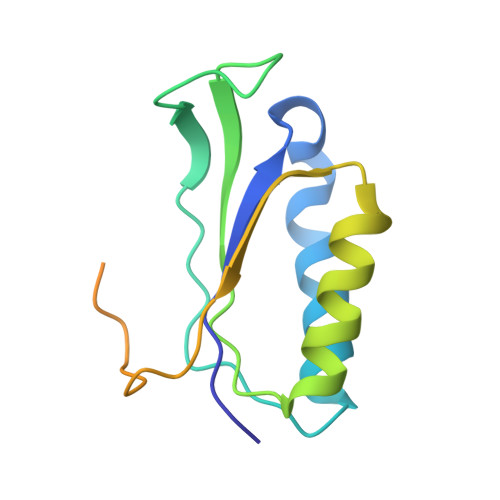
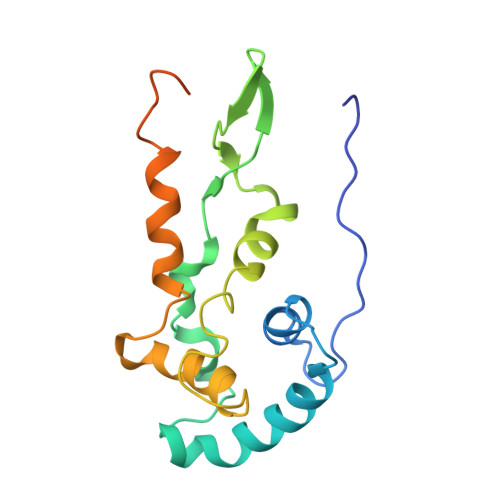
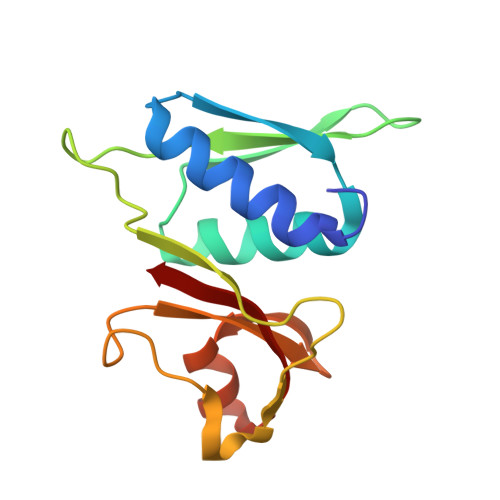
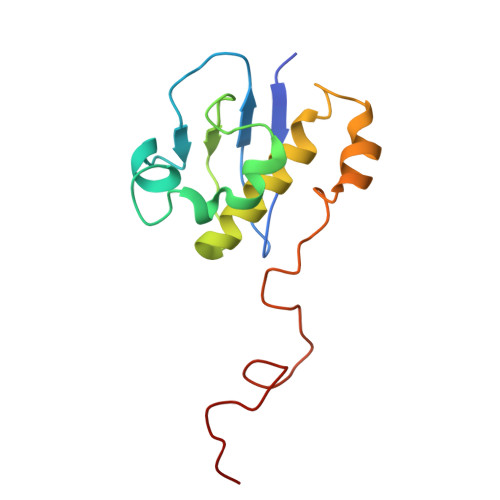
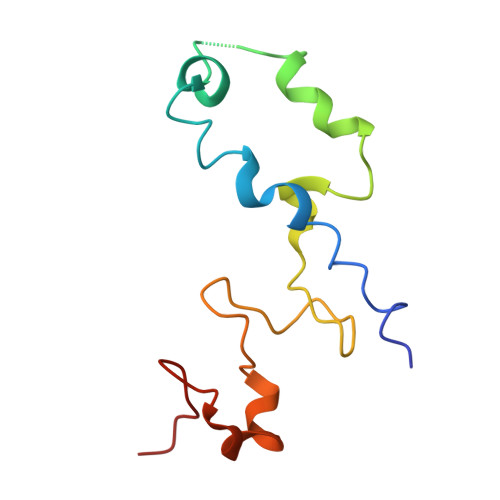
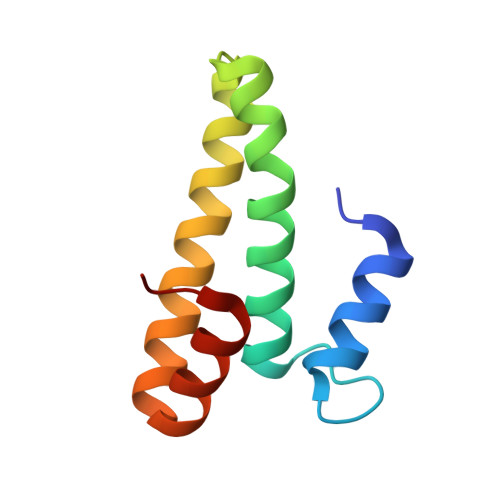
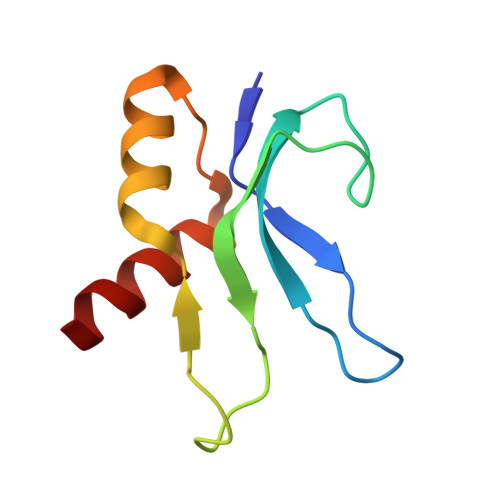
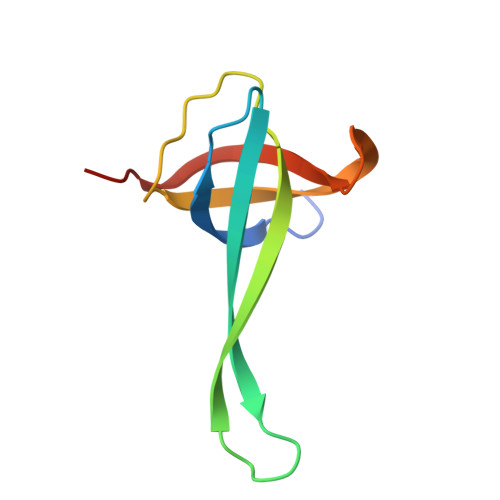
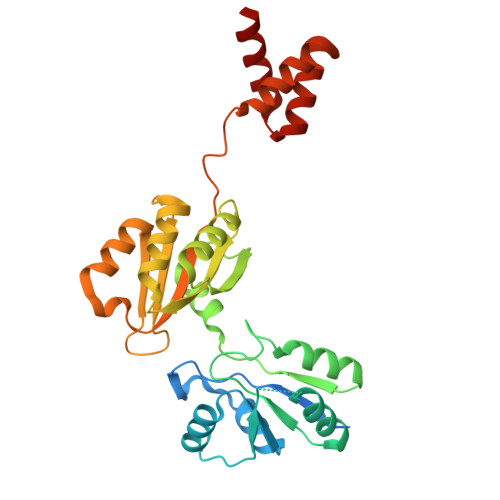
Mechanism of 30S subunit recognition and modification by the conserved bacterial ribosomal RNA methyltransferase RsmI.
Barmada, M.I., McGinity, E.N., Nandi, S., Dey, D., Zelinskaya, N., Harris, G.M., Comstock, L.R., Dunham, C.M., Conn, G.L.(2025) bioRxiv
- PubMed: 40909654
- DOI: https://doi.org/10.1101/2025.08.28.672957
- Primary Citation of Related Structures:
9PZG - PubMed Abstract:
Ribosomal RNA (rRNA) modifications are important for ribosome function and can influence bacterial susceptibility to ribosome-targeting antibiotics. The universally conserved 16S rRNA nucleotide C1402, for example, is the only 2'- O -methylated nucleotide in the bacterial small (30S) ribosomal subunit and this modification fine tunes the shape and structure of the peptidyl tRNA binding site. The Cm1402 modification is incorporated by the conserved bacterial 16S rRNA methyltransferase RsmI, but it is unclear how RsmI is able to recognize its 30S substrate and specifically modify its buried target nucleotide. We determined a 2.42 Å resolution cryo-EM structure of the RsmI-30S complex and, with accompanying functional analyses, show that RsmI anchors itself to the 30S subunit through multiple contacts with a conserved 16S rRNA tertiary surface present only in the assembled subunit. This positions RsmI to induce an extensive h44 distortion to access C1402 that is unprecedented among 16S rRNA methyltransferases characterized to date. These analyses also reveal an essential contribution to 30S subunit interaction made by the previously structurally uncharacterized RsmI C-terminal domain, RsmI-induced RNA-RNA interactions with C1402, and an unappreciated dependence on a divalent metal ion for catalysis, marking RsmI as the first of a distinct class of metal- and SAM-dependent RNA O -methyltransferases. This study significantly expands our mechanistic understanding of how intrinsic bacterial methyltransferases like RsmI modify their rRNA targets. Further, recognition of distant ribosome features and extensive unfolding of a critical rRNA functional center point to a potential role in accurate 30S subunit biogenesis.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, Georgia, 30322, USA.
Organizational Affiliation: