Co-crystal structure of two CCM2 PTB domains bound to a KRIT1 peptide encompassing NPxF2 and NPxF3
Fisher, O.S., Boggon, T.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Malcavernin | A, B, C, F [auth D] | 180 | Homo sapiens | Mutation(s): 0 Gene Names: CCM2, C7orf22, PP10187 | 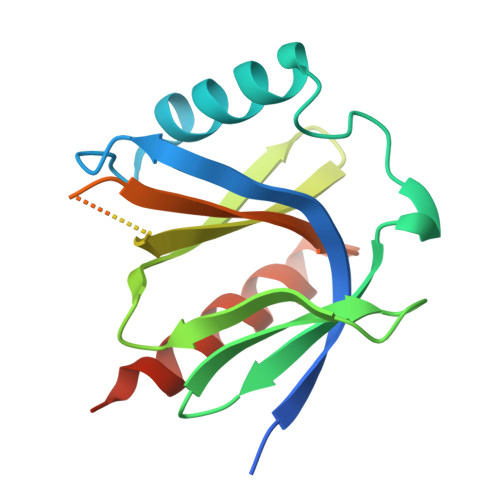 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9BSQ5 (Homo sapiens) Explore Q9BSQ5 Go to UniProtKB: Q9BSQ5 | |||||
PHAROS: Q9BSQ5 GTEx: ENSG00000136280 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9BSQ5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Krev interaction trapped protein 1 | D [auth E], E [auth F] | 29 | Homo sapiens | Mutation(s): 0 | 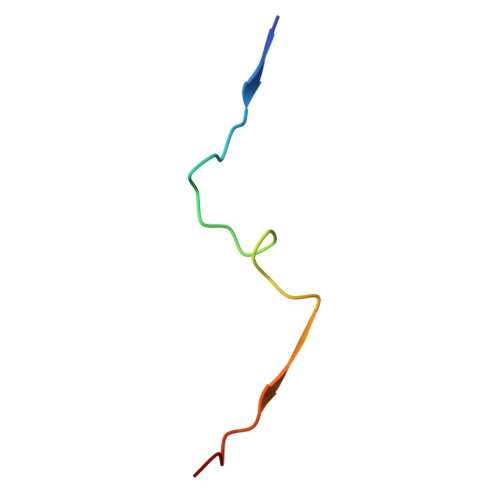 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O00522 (Homo sapiens) Explore O00522 Go to UniProtKB: O00522 | |||||
PHAROS: O00522 GTEx: ENSG00000001631 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O00522 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 45.198 | α = 90 |
| b = 120.441 | β = 102.73 |
| c = 67.778 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of Neurological Disorders and Stroke (NIH/NINDS) | United States | R01NS134606 |
| American Heart Association | United States | 961309 |