Discovery of PF-07054894, a Potent Squaramide-Based CCR6 Antagonist Displaying High CXCR2 Selectivity.
Schnute, M.E., Wu, H., Trujillo, J.I., Li, W., Wasilko, D.J., Alley, J., Arnold, E.P., Bagley, S.W., Berstein, G., Blakemore, D.C., Bundesmann, M.W., Chinigo, G.M., Choi, C., Cooke, R., Crouse, K., Dermenci, A., Dickinson, T., Flick, A., Frisbie, R.K., Garcia-Irizarry, C., Gerstenberger, B.S., Hepworth, D., Kaila, N., Kung, D.W., Lamb, D., Lavergne, S.Y., Limburg, D.C., Liu, S., Lombardo, V.M., Mondal, P.K., Mousseau, J.J., Narayanan, A., Niljianskul, N., Nuhant, P., Pan, S., Primiano, M.J., Rappas, M., Schmitt, D.C., Steyn, S.J., Unwalla, R., Vasa, D., Vazquez, M.L., Vincent, F., Yang, X., Thorarensen, A.(2025) J Med Chem 68: 20689-20716
- PubMed: 40977184
- DOI: https://doi.org/10.1021/acs.jmedchem.5c01946
- Primary Citation of Related Structures:
9PEE - PubMed Abstract:
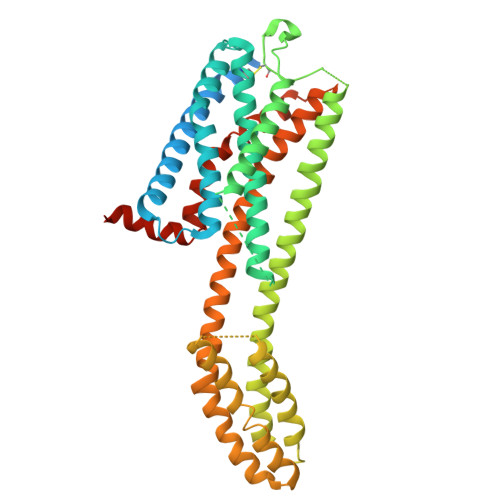
The G protein-coupled receptor (GPCR) CCR6 mediates the migration of pathogenic immune cells to the site of inflammation in response to the chemokine CCL20. CCR6 is an attractive target for the treatment of chronic autoimmune disease as antagonism of the receptor is expected to block CCL20-mediated immune cell recruitment. High-throughput-screening identified a squaramide-based, allosteric antagonist of CCR6 that potently inhibited CCL20-mediated T cell chemotaxis, but it also inhibited CXCL1-mediated neutrophil chemotaxis through CXCR2 antagonism. Lead optimization achieved differentiation from CXCR2 through identification of a methyl substituent-dependent selectivity switch or "magic methyl for selectivity". The mechanism for this effect is rooted in different antagonist-induced ligand-receptor behaviors, insurmountable for CCR6 and surmountable for CXCR2. Herein, we report the discovery and preclinical evaluation of the CCR6 antagonist PF-07054894 ( 18l ) which has advanced to clinical trials as a first in class approach targeting the receptor.
- Medicine Design, Pfizer Inc, Cambridge, Massachusetts 02139, United States.
Organizational Affiliation: