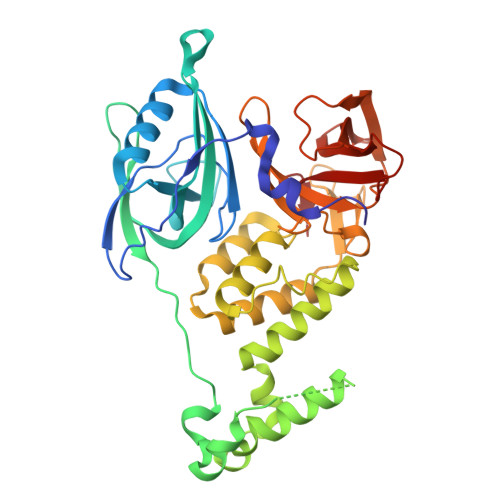
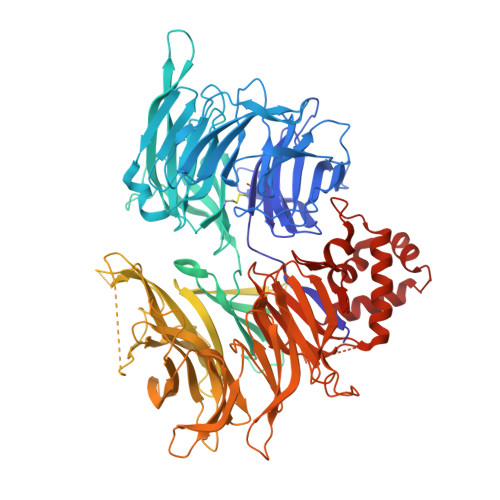
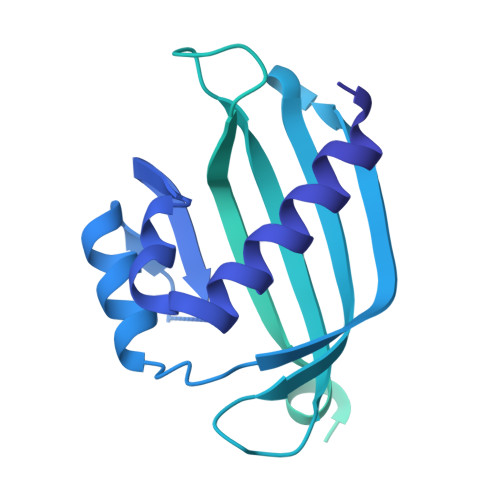
Cereblon induces G3BP2 neosubstrate degradation using molecular surface mimicry
Annunziato, S., Quan, C., Donckele, E.J., Lamberto, I., Bunker, R.D., Zlotosch, M., Schwander, L., Murthy, A., Wiedmer, L., Staehly, C., Matysik, M., Gilberto, S., Kapsitidou, D., Wible, D., de Donatis, G.D., Trenh, P., SriRamaratnam, R., Strande, V., Almeida, R., Dolgikh, E., DeMarco, B., Tsai, J., Sadok, A., Zarayskiy, V., Walter, M., Tiedt, R., Lumb, K.J., Bonenfant, D., Fasching, B., Castle, J.C., Townson, S.A., Petzold, G., Gainza, P.(2026) Nat Struct Mol Biol