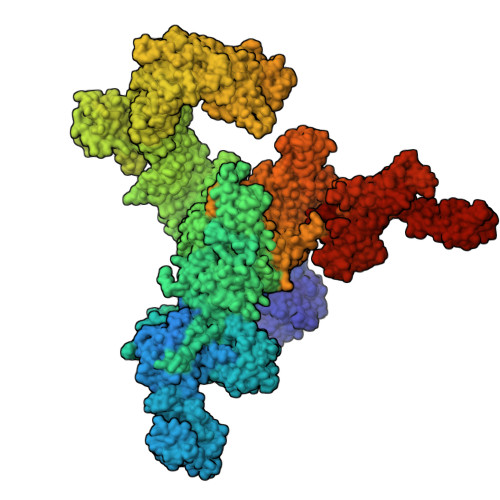
Cryo-electron microscopy reveals sequential binding and activation of Ryanodine Receptors by statin triplets.
Molinarolo, S., Valdivia, C.R., Valdivia, H.H., Van Petegem, F.(2025) Nat Commun 16: 11508-11508
- PubMed: 41266329
- DOI: https://doi.org/10.1038/s41467-025-66522-0
- Primary Citation of Related Structures:
9OL4, 9OL5, 9OL6, 9PWO - PubMed Abstract:
Statins are the most prescribed class of drugs and inhibit a key enzyme in the cholesterol biosynthesis pathway. Many patients have reported mild to severe muscle related symptoms and a subset are at risk for rhabdomyolysis. Sequence variants in RyR1, the skeletal muscle Ryanodine Receptor, correlate with intolerance to statins, but whether RyR1 can bind statins directly has remained unclear. Here we report cryo-EM structures of RyR1 in the absence and presence of atorvastatin, firmly establishing RyR1 as an unintended off-target. Our results show an unusual binding mode whereby three atorvastatin molecules bind together in a cleft formed by the pseudo-voltage sensing domain, making extensive interactions with each other and with RyR1. Atorvastatin activates RyR1 in a sequential way, whereby one statin per subunit can bind to the transmembrane region of a closed RyR1, with small structural perturbations that prime the channel for opening. Binding of two additional statins per subunit is associated with a widening of the pseudo-voltage sensing domain that triggers opening of the pore. Comparison with atorvastatin binding to HMG-CoA reductase, its intended target, offers clues on how to modify the statin to reduce RyR1 binding, while leaving binding to HMG-CoA reductase unperturbed.
- Department of Biochemistry and Molecular Biology, The Life Sciences Institute, University of British Columbia, Vancouver, British Columbia, Canada.
Organizational Affiliation: