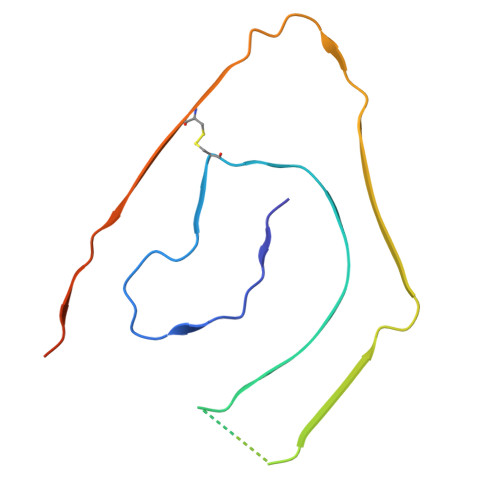
Cryo-EM of Cardiac AL-224L Amyloid Reveals Shared Structural Motifs and Mutation-induced Differences in lambda 6 Light Chain Fibrils.
Hicks, C.W., Prokaeva, T., Spencer, B., Jayaraman, S., Huda, N., Wong, S., Chen, H., Sanchorawala, V., Lavatelli, F., Gursky, O.(2025) J Mol Biology 438: 169591-169591
- PubMed: 41389958
- DOI: https://doi.org/10.1016/j.jmb.2025.169591
- Primary Citation of Related Structures:
9OKA - PubMed Abstract:
In light chain amyloidosis (AL), aberrant monoclonal antibody light chains (LCs) deposit in vital organs causing organ damage. Each AL patient features a unique LC; previous cryogenic electron microscopy (cryo-EM) studies revealed different amyloid structures in different AL patients. How LC mutations influence amyloid structures remains unclear. We report a cryo-EM structure of cardiac AL-224L amyloid (2.92 Å resolution) from λ6-LC family, which is overrepresented in AL amyloidosis. Comparison with λ6-LC structures from two other patients reveals similarities in amyloid folds, along with major differences caused by specific mutations. Differences in AL-224L include altered C-terminal conformation with an exposed surface forming an apparent ligand-binding site; an enlarged hydrophilic pore with orphan density; and altered steric zipper registry with backbone flipping, which likely represent general adaptive mechanisms in amyloids. The results reveal shared features in λ6-LC amyloid folds and suggest how mutation-induced structural changes influence amyloid-ligand interactions in a patient-specific manner.
- Department of Pharmacology, Physiology & Biophysics, Chobanian & Avedisian School of Medicine, Boston University, Boston, MA, USA. Electronic address: hickscw@bu.edu.
Organizational Affiliation: