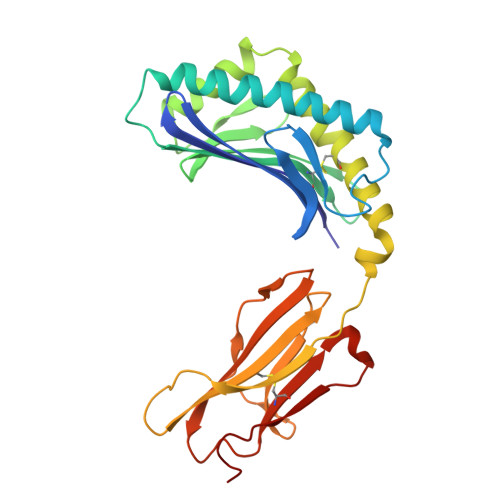
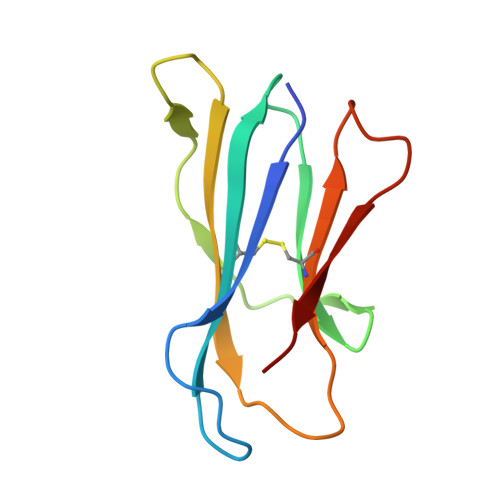
Sideways lipid presentation by the antigen-presenting molecule CD1c.
Cao, T.P., Liao, G.R., Cheng, T.Y., Chen, Y., Ciacchi, L., Fulford, T.S., Farquhar, R., Kollmorgen, J., Mayfield, J.A., Uldrich, A.P., Ng, E.Z.Q., Ogg, G.S., Godfrey, D.I., Gherardin, N.A., Chen, Y.L., Moody, D.B., Shahine, A., Rossjohn, J.(2025) Nat Commun
- PubMed: 41476042
- DOI: https://doi.org/10.1038/s41467-025-67732-2
- Primary Citation of Related Structures:
9OHT, 9OHU, 9OHV, 9OHW, 9OHX, 9OHY, 9OHZ, 9OI0 - PubMed Abstract:
For the MHC, MR1 and CD1 systems, antigen recognition involves contact of the membrane distal surfaces of both the αβ T cell receptor (TCR) and the antigen-presenting molecule. Whether other antigen display mechanisms by antigen-presenting molecules operate remains unknown. Here, we report mass spectrometry analyses of endogenous lipids captured by CD1c when bound to an autoreactive αβ TCR. CD1c binds twenty-six lipid species with bulky headgroups that cannot fit within the tight TCR-CD1c interface. We determined the crystal structures of CD1c presenting several gangliosides, revealing a general mechanism whereby two lipids, rather than one, are bound in the CD1c cleft. Bulky lipids are oriented sideways so that their polar headgroups protrude laterally through a side portal of the CD1c molecule - an evolutionarily conserved structural feature. The sideways-presented ganglioside headgroups do not hinder TCR binding and so represent a mechanism that allows autoreactive TCR recognition of CD1c. In addition, ex vivo studies showed that the sideways-presented gangliosides can also represent TCR recognition determinants. These findings reveal that CD1c simultaneously presents two lipid antigens from the top and side of its cleft, a general mechanism that differs markedly from other antigen-presenting molecules.
- Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, VIC, Australia.
Organizational Affiliation: