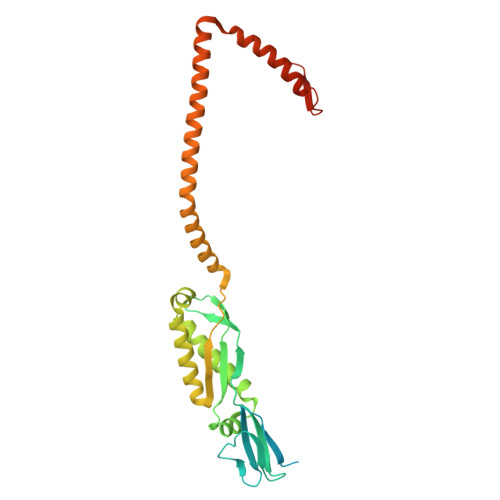
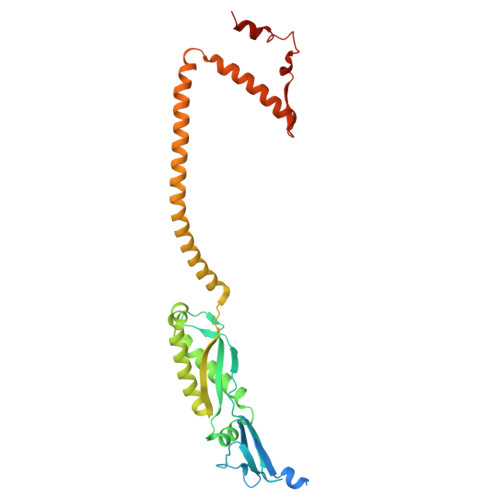
Structures of human organellar SPFH protein complexes.
Gao, J., Sherpa, D., Kupko, N., Chino, H., Zeng, J., Shao, S.(2025) Nat Commun 16: 10064-10064
- PubMed: 41249155
- DOI: https://doi.org/10.1038/s41467-025-65078-3
- Primary Citation of Related Structures:
9O9U, 9O9Z, 9OA0 - PubMed Abstract:
Stomatin, Prohibitin, Flotillin, and HflK/C (SPFH) family proteins are found in all kingdoms of life and in multiple eukaryotic organelles. SPFH proteins assemble into homo- or hetero-oligomeric rings that form domed structures. Most SPFH assemblies also abut a cellular membrane, where they are implicated in diverse functions ranging from membrane organization to protein quality control. However, the precise architectures of different SPFH complexes remain unclear. Here, we report single-particle cryo-EM structures of the endoplasmic reticulum (ER)-resident Erlin1/2 complex and the mitochondrial prohibitin (PHB1/2) complex, revealing assemblies of 13 heterodimers of Erlin1 and Erlin2 and 11 heterodimers of PHB1 and PHB2, respectively. We also describe key interactions underlying the architecture of each complex and conformational heterogeneity of the PHB1/2 complex. Our findings elucidate the distinct stoichiometries and properties of human organellar SPFH complexes and highlight common principles of SPFH complex organization.
- Department of Cell Biology, Harvard Medical School, Boston, MA, US.
Organizational Affiliation: