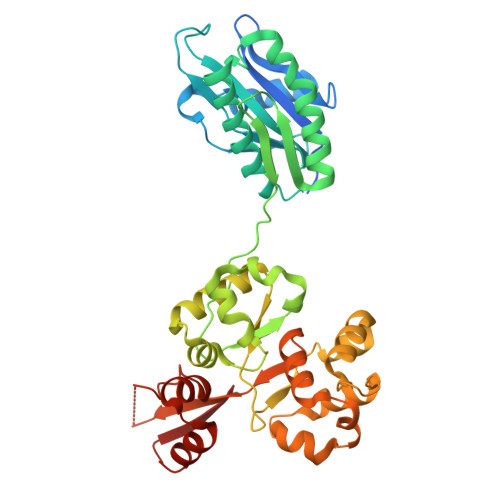
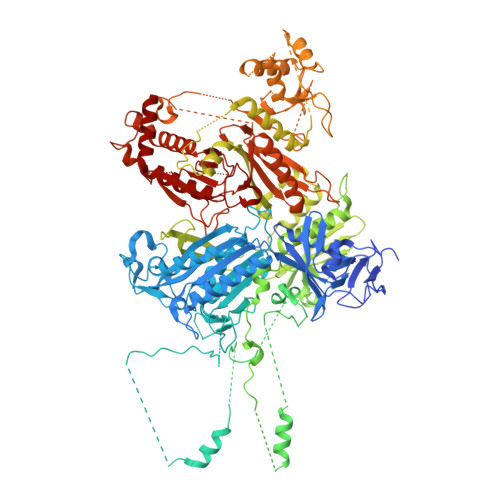
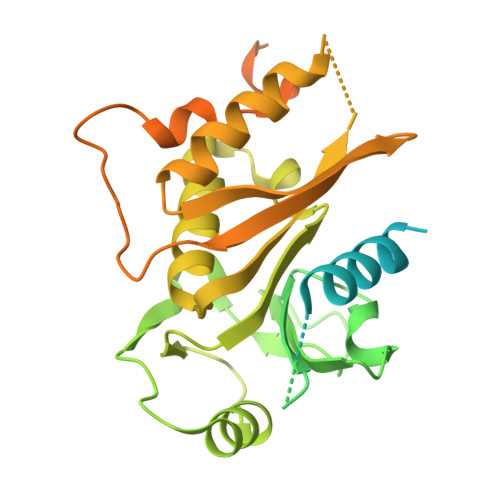
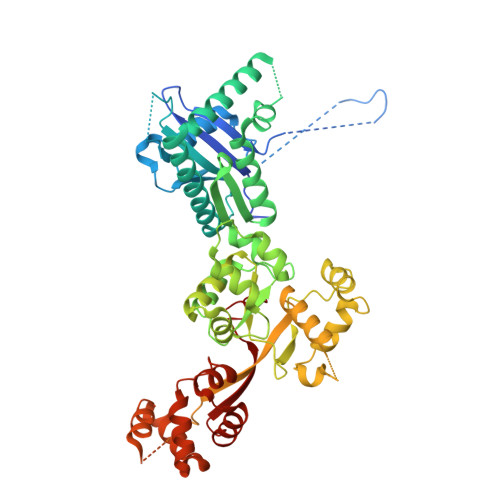
Structure of the lysosomal KICSTOR-GATOR1-SAMTOR nutrient-sensing supercomplex.
Lupton, C.J., Bayly-Jones, C., Dong, S., Lam, T., Luo, W., Jones, G.D., Mastos, C., Frescher, N.J., Lim, S.S., Keen, A.C., Formosa, L.E., Venugopal, H., Chang, Y.G., Halls, M.L., Ellisdon, A.M.(2026) Cell 189: 1185-1200.e28
- PubMed: 41512879
- DOI: https://doi.org/10.1016/j.cell.2025.12.005
- Primary Citation of Related Structures:
9O5A, 9O5D, 9O5E - PubMed Abstract:
The guanosine triphosphate (GTP)-bound state of the heterodimeric Rag GTPases functions as a molecular switch regulating mechanistic target of rapamycin complex 1 (mTORC1) activation at the lysosome downstream of amino acid fluctuations. Under low amino acid conditions, GTPase-activating protein (GAP) activity toward Rags 1 (GATOR1) promotes RagA GTP hydrolysis, preventing mTORC1 activation. KICSTOR recruits and regulates GATOR1 at the lysosome by undefined mechanisms. Here, we resolve the KICSTOR-GATOR1 structure, revealing a striking ∼60-nm crescent-shaped assembly. GATOR1 anchors to KICSTOR via an extensive interface, and mutations that disrupt this interaction impair mTORC1 regulation. The S-adenosylmethionine sensor SAMTOR binds KICSTOR in a manner incompatible with metabolite binding, providing structural insight into methionine sensing via SAMTOR-KICSTOR association. We discover that KICSTOR and GATOR1 form a dimeric supercomplex. This assembly restricts GATOR1 to an orientation that favors the low-affinity active GAP mode of Rag GTPase engagement while sterically restricting access to the high-affinity inhibitory mode, consistent with a model of an active lysosomal GATOR1 docking complex.
- Cancer Program, Biomedicine Discovery Institute, Monash University, Clayton 3800, VIC, Australia.
Organizational Affiliation: