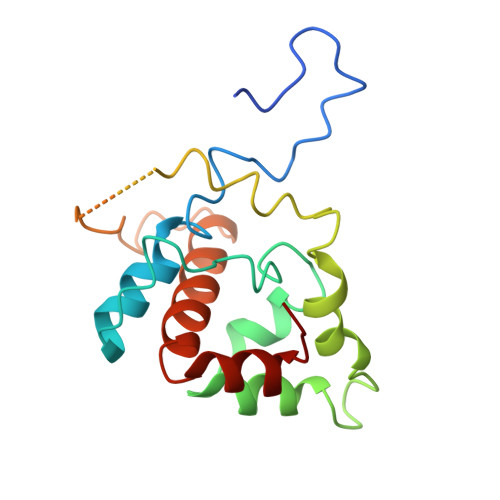
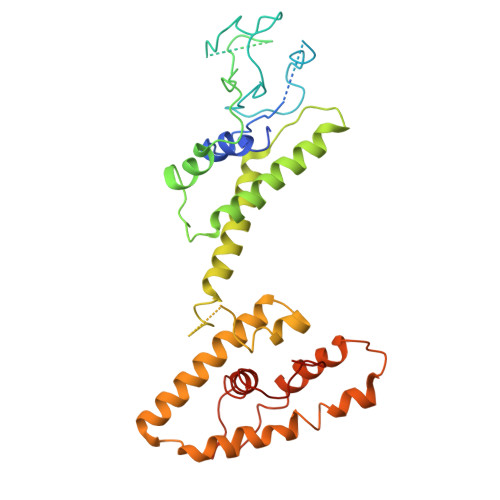
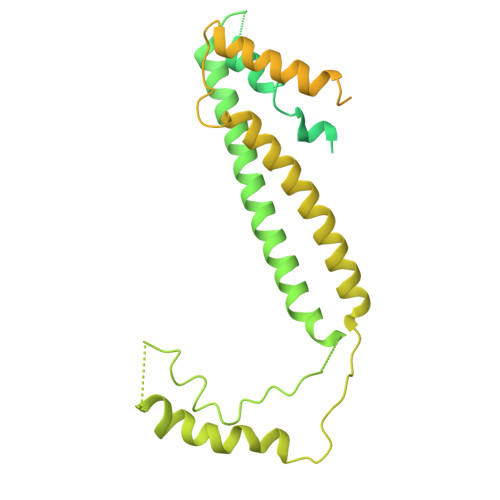
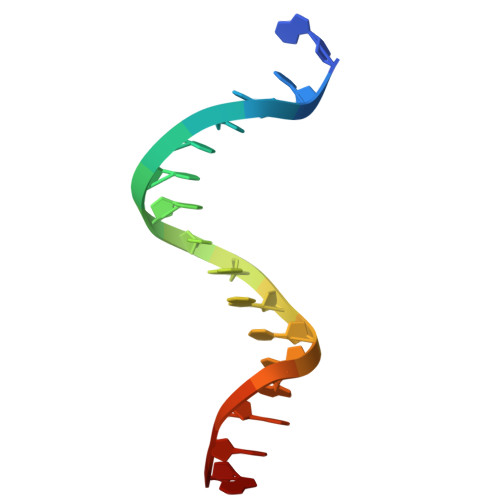
Structural basis for Lamassu-based antiviral immunity and its evolution from DNA repair machinery.
Haudiquet, M., Chakravarti, A., Zhang, Z., Ramirez, J.L., Herrero Del Valle, A., Olinares, P.D.B., Lavenir, R., Ahmed, M.A., de la Cruz, M.J., Chait, B.T., Sternberg, S.H., Bernheim, A., Patel, D.J.(2025) Proc Natl Acad Sci U S A 122: e2519643122-e2519643122
- PubMed: 41252147
- DOI: https://doi.org/10.1073/pnas.2519643122
- Primary Citation of Related Structures:
9NXX, 9NY1, 9NY5, 9NYG - PubMed Abstract:
Bacterial immune systems exhibit remarkable diversity and modularity, as a consequence of the continuous selective pressures imposed by phage predation. Despite recent mechanistic advances, the evolutionary origins of many antiphage immune systems remain elusive, especially for those that encode homologs of the structural maintenance of chromosomes (SMC) superfamily, which are essential for chromosome maintenance and DNA repair across domains of life. Here, we elucidate the structural basis and evolutionary emergence of Lamassu, a bacterial immune system family featuring diverse effectors but a core conserved SMC-like sensor. Using cryo-EM, we determined structures of the Vibrio cholerae Lamassu complex in both apo- and dsDNA-bound states, revealing unexpected stoichiometry and topological architectures. We further demonstrate how Lamassu specifically senses dsDNA ends in vitro and phage replication origins in vivo, thereby triggering the formation of LmuA tetramers that activate its Cap4 nuclease domain. Our findings reveal that Lamassu evolved via exaptation of the bacterial Rad50-Mre11 DNA repair system to form a compact, modular sensor for viral replication, exemplifying how cellular machinery can be co-opted for novel immune functions.
- Institut Pasteur, Université Paris-Cité, CNRS UMR2535, Department of Genomes and Genetics, Molecular Diversity of Microbes, Paris 75015, France.
Organizational Affiliation: