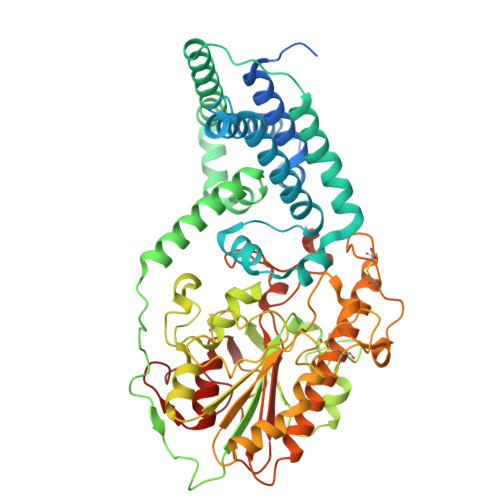
Mechanistic basis of antimicrobial resistance mediated by the phosphoethanolamine transferase MCR-1.
Zinkle, A.P., Batista, M.B., Herrera, C.M., Erramilli, S.K., Kloss, B., Ashraf, K.U., Nosol, K., Zhang, G., Cater, R.J., Marty, M.T., Kossiakoff, A.A., Trent, M.S., Nygaard, R., Stansfeld, P.J., Mancia, F.(2025) Nat Commun 16: 10516-10516
- PubMed: 41298376
- DOI: https://doi.org/10.1038/s41467-025-65515-3
- Primary Citation of Related Structures:
9NWW - PubMed Abstract:
Polymyxins are used to treat infections caused by multidrug-resistant Gram-negative bacteria. They are cationic peptides that target the negatively charged lipid A component of lipopolysaccharides, disrupting the outer membrane and lysing the cell. Polymyxin resistance is conferred by inner-membrane enzymes, such as phosphoethanolamine transferases, which add positively charged phosphoethanolamine to lipid A. Here, we present the structure of MCR-1, a plasmid-encoded phosphoethanolamine transferase, in its liganded form. The phosphatidylethanolamine donor substrate is bound near the active site in the periplasmic domain, and lipid A is bound over 20 Å away, within the transmembrane region. Integrating structural, biochemical, and drug-resistance data with computational analyses, we propose a two-state model in which the periplasmic domain rotates to bring the active site to lipid A, near the preferential phosphate modification site for MCR-1. This enzymatic mechanism may be generally applicable to other phosphoform transferases with large, globular soluble domains.
- Department of Physiology and Cellular Biophysics, Columbia University Irving Medical Center, New York, NY, USA.
Organizational Affiliation: