Structure of MurJ in complex with single gene lysis protein from phage PP7
Li, Y.E., Clemons, W.M.To be published.
Experimental Data Snapshot
Starting Models: experimental, in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lipid II flippase MurJ | 614 | Escherichia coli K-12 | Mutation(s): 3 Gene Names: murJ, mviN, yceN, b1069, JW1056 | 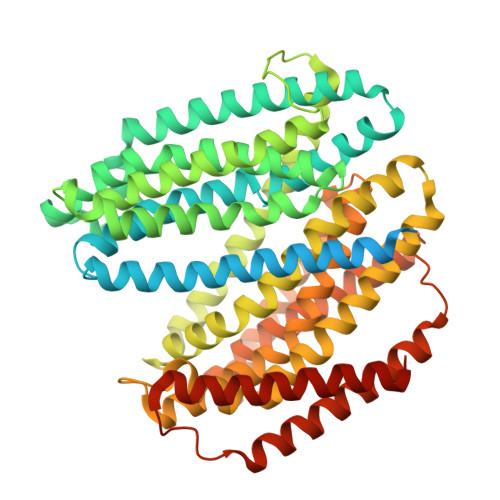 | |
UniProt | |||||
Find proteins for P0AF16 (Escherichia coli (strain K12)) Explore P0AF16 Go to UniProtKB: P0AF16 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AF16 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lysis protein | 62 | Pseudomonas phage PP7 | Mutation(s): 3 | 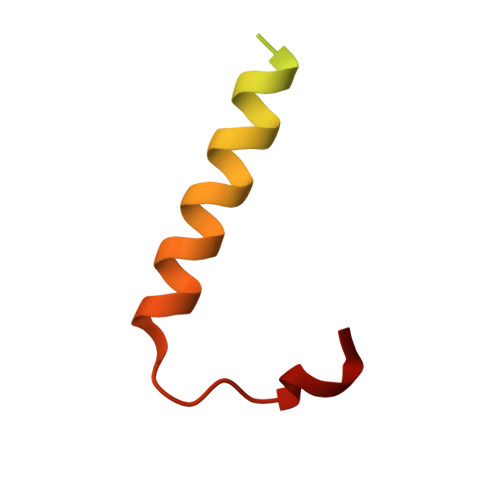 | |
UniProt | |||||
Find proteins for Q38063 (Pseudomonas phage PP7) Explore Q38063 Go to UniProtKB: Q38063 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q38063 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 4.5.3 |
| MODEL REFINEMENT | PHENIX | 1.21_5207 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM114611 |
| The G. Harold and Leila Y. Mathers Foundation | United States | to W.M.C. |