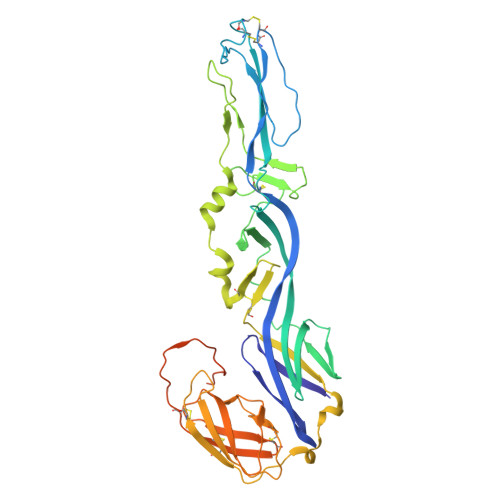
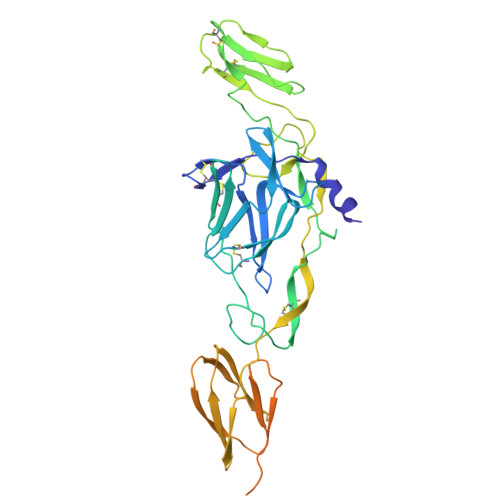
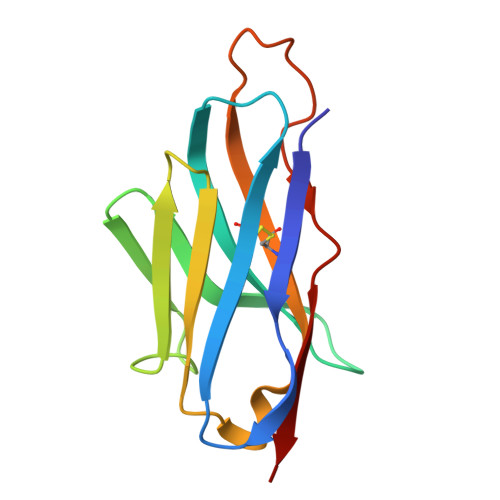
Demonstration of in vivo efficacy, cryo-EM-epitope identification, and breadth of two anti-alphavirus bispecific single domain antibodies.
Gardner, C.L., Pletnev, S., Liu, J.L., Anderson, G.P., Shriver-Lake, L.C., Bylund, T., Green, C., Stephens, T., Sutton, M.S., Tsybovsky, Y., Roederer, M., Kwong, P.D., Zhou, T., Goldman, E.R., Burke, C.W.(2026) J Virol 100: e0187525-e0187525
- PubMed: 41400357
- DOI: https://doi.org/10.1128/jvi.01875-25
- Primary Citation of Related Structures:
9NRX, 9NRY, 9NRZ - PubMed Abstract:
Venezuelan equine encephalitis virus (VEEV) is an arbovirus that causes a disease in which 4%-14% of individuals can develop neurological symptoms. Prior to 1970, VEEV was developed as a biological threat agent due to its stability and high morbidity when administered by aerosol. Currently, no FDA-licensed vaccines nor therapeutics for VEEV exist. Single-domain antibodies (sdAbs) may provide a therapeutic option due to their small size and ability to bind recessed epitopes not recognized by conventional antibodies. This study identified two bivalent sdAbs that were able to protect mice from a lethal challenge against both epizootic and enzootic subtypes of VEEV. Cryo-EM structures of sdAb-VEEV complexes revealed the sdAbs that comprised the bivalent sdAbs to recognize a mixture of conserved and non-conserved regions of the VEEV envelope proteins. While all three of the cryo-EM-characterized epitopes were unique in terms of their recognized VEEV residues, two sdAbs, V2B3 and V2C3, overlapped sterically, explaining why only their combinations with the non-sterically overlapping sdAb V3A8f, which composed the bivalent sdAbs described here, were so particularly effective. Binding and neutralization studies found that the bivalent sdAbs have the potential to be broad-spectrum anti-alphavirus therapeutics as they cross-neutralize multiple alphaviruses.IMPORTANCEAlphaviruses are no longer geographically constrained to one region of the world but are expanding to be of global concern. In many regions of the world, multiple alphaviruses co-circulate; therefore, having a therapeutic that is pan-alphavirus is important. A cocktail of multiple pan-alphavirus binding/neutralizing antibodies (Abs) may provide optimal coverage against alphaviruses while decreasing the prevalence of viral escape mutants, which could cause the therapeutic to no longer be efficacious. Structures of these Abs, defining their recognition, could assist in identifying optimal combinations. A bivalent pan-alphavirus single-domain antibody could be used in a cocktail with already identified alphavirus IgG antibodies.
- Virology Division, United States Army Medical Research Institute of Infectious Diseases (USAMRIID), Fort Detrick, Frederick, Maryland, USA.
Organizational Affiliation: