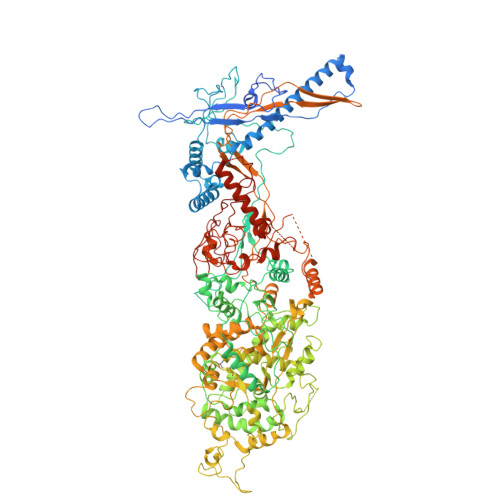
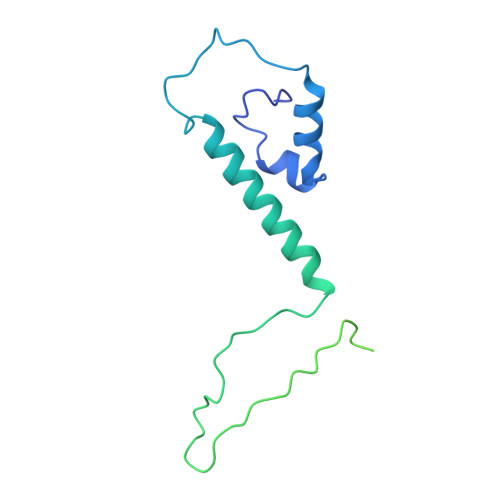
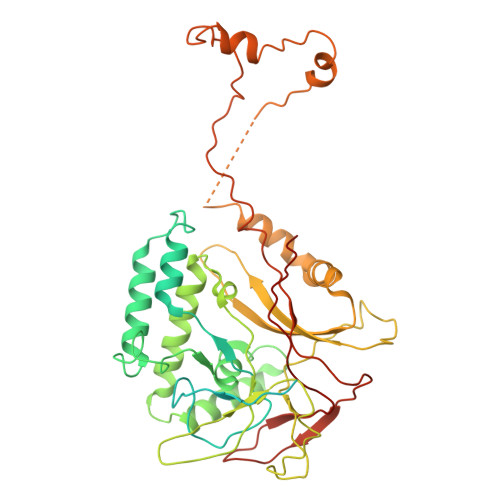
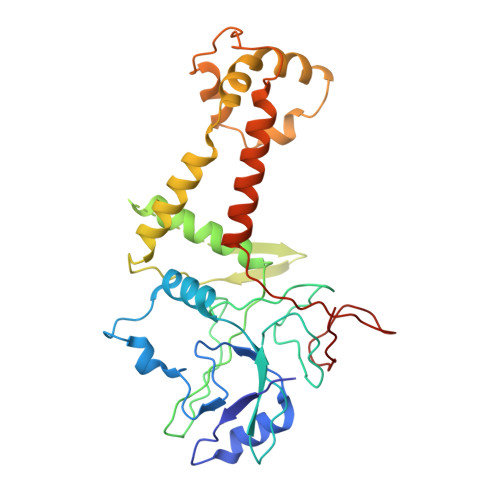
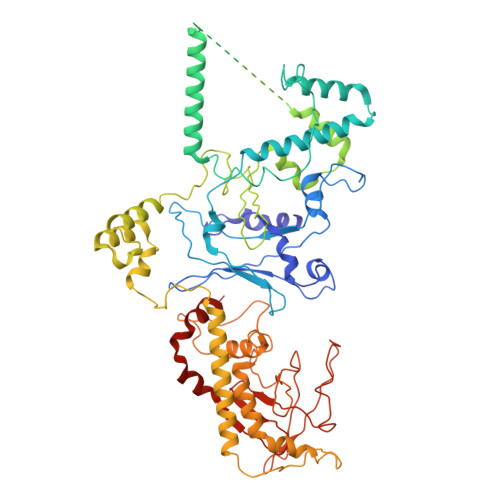
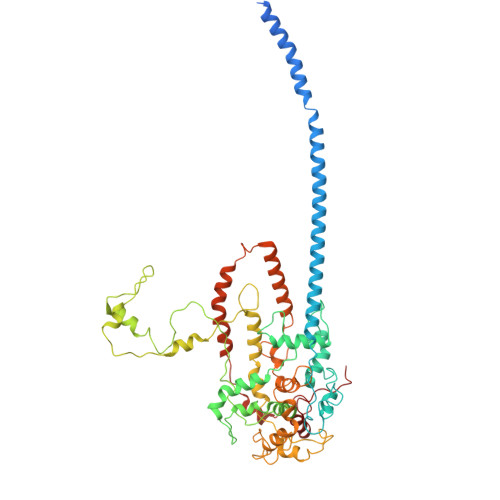
Cryogenic Electron Tomography Redefines Herpesvirus Capsid Assembly Intermediates Inside the Cell Nucleus.
Oliver, S.L., Chen, M., Engel, L., Hecksel, C.W., Zhou, X., Schmid, M.F., Arvin, A.M., Chiu, W.(2025) bioRxiv
- PubMed: 40666836
- DOI: https://doi.org/10.1101/2025.06.27.661840
- Primary Citation of Related Structures:
9NO1 - PubMed Abstract:
Herpesviruses encapsulate their double-stranded DNA (dsDNA) genomes within an icosahedral nucleocapsid formed in the infected cell nucleus. Four biochemically purified nucleocapsids have been characterized, but their roles in herpesvirus replication remain controversial. The status of the capsid vertex-specific component (CVSC), essential for capsid stability and dsDNA packaging and retention, is also unclear. By integrating cryogenic focused ion beam milling with electron tomography and subtomogram averaging, we derived atomic models for all protein components, including the CVSC, across different herpesvirus capsid types within infected cell nuclei. Focused classification of pentonal vertex densities revealed differences in CVSC occupancy between genome-filled capsids and capsids lacking dsDNA, highlighting structural heterogeneity and providing insight into distinct capsid assembly stages in situ . These intra-nuclear findings redefine the maturation model of herpesvirus capsid assembly, advancing the understanding of herpesvirus replication, and demonstrate the effectiveness of in situ electron imaging by studying virus assembly within host cells.