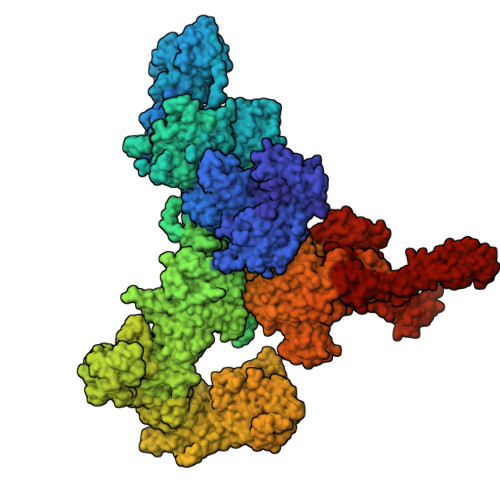
Structural basis for simvastatin-induced skeletal muscle weakness associated with type 1 ryanodine receptor T4709M mutation.
Weninger, G., Dridi, H., Reiken, S., Yuan, Q., Zhao, N., Groom, L., Leigh, J., Liu, Y., Tchagou, C., Kang, J., Chang, A., Luna-Figueroa, E., Miotto, M.C., Wronska, A., Dirksen, R.T., Marks, A.R.(2025) J Clin Invest 135
- PubMed: 41392983
- DOI: https://doi.org/10.1172/JCI194490
- Primary Citation of Related Structures:
9NMN, 9NMO, 9NMP, 9NMQ, 9NMR - PubMed Abstract:
Statins lower cholesterol, reducing the risk of heart disease, and are among the most frequently prescribed drugs. Approximately 10% of individuals develop statin-associated muscle symptoms (SAMS; myalgias, rhabdomyolysis, and muscle weakness), often rendering them statin intolerant. The mechanism underlying SAMS remains poorly understood. Patients with mutations in the skeletal muscle ryanodine receptor 1 (RyR1)/calcium release channel can be particularly intolerant of statins. High-resolution structures revealed simvastatin binding sites in the pore region of RyR1. Simvastatin stabilized the open conformation of the pore and activated the RyR1 channel. In a mouse expressing a mutant RyR1-T4709M found in a patient with profound statin intolerance, simvastatin caused muscle weakness associated with leaky RyR1 channels. Cotreatment with a Rycal drug that stabilizes the channel closed state prevented simvastatin-induced muscle weakness. Thus, statin binding to RyR1 can cause SAMS, and patients with RyR1 mutations may represent a high-risk group for statin intolerance.
- Department of Physiology and Cellular Biophysics, Clyde and Helen Wu Center for Molecular Cardiology, Vagelos College of Physicians and Surgeons, Columbia University Medical Center, New York, New York, USA.
Organizational Affiliation: