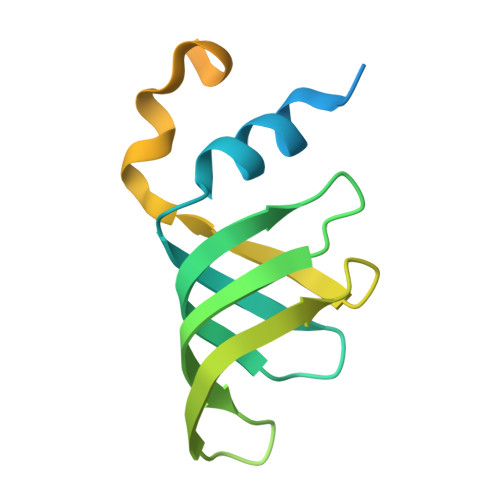
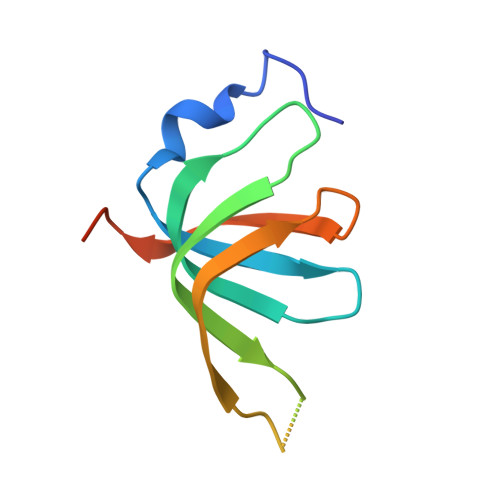
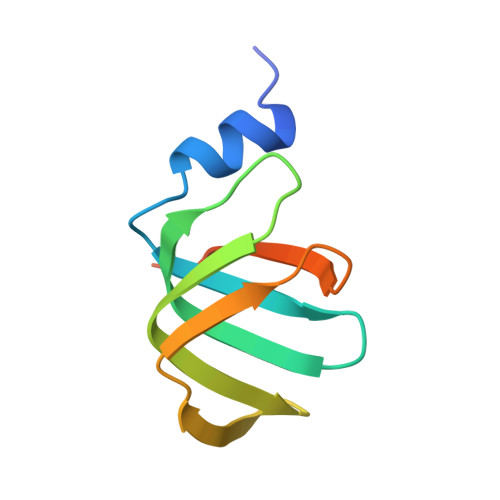
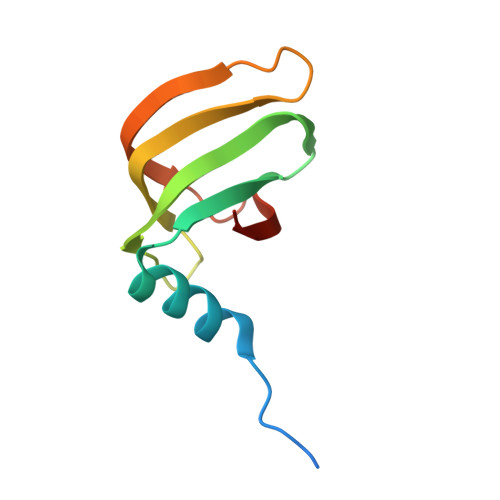
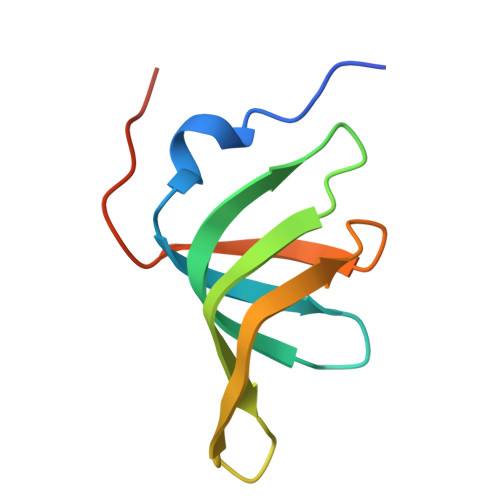
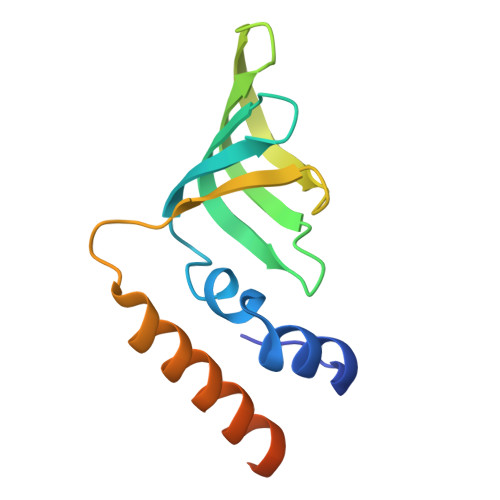
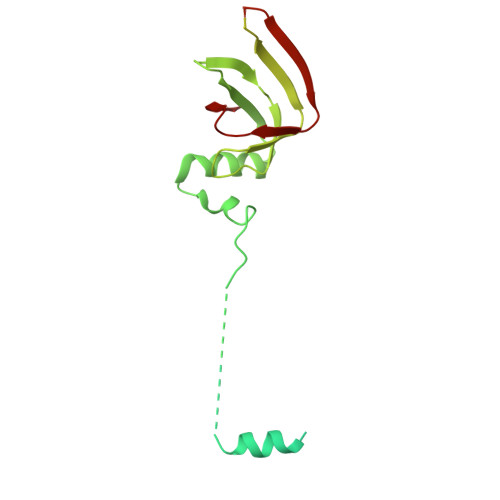
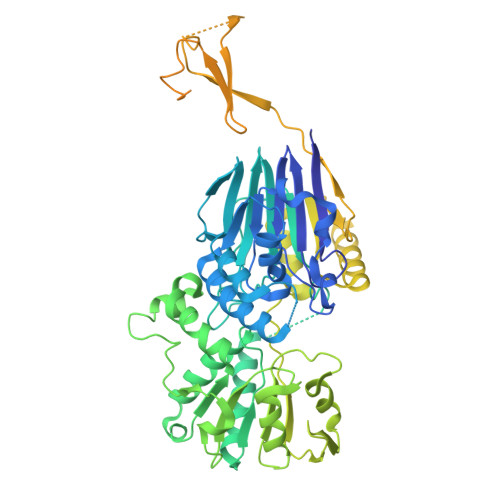
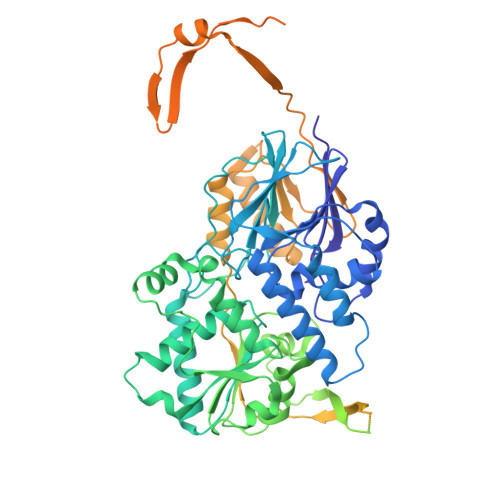
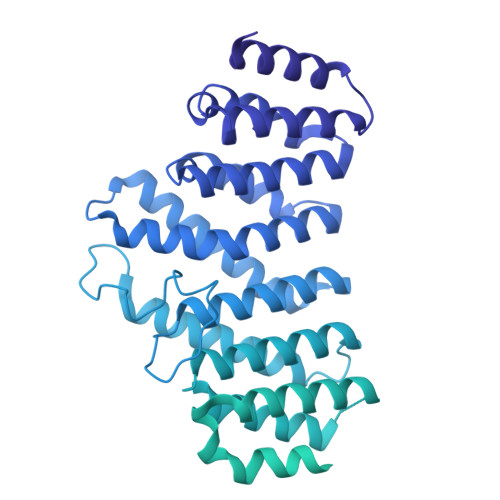
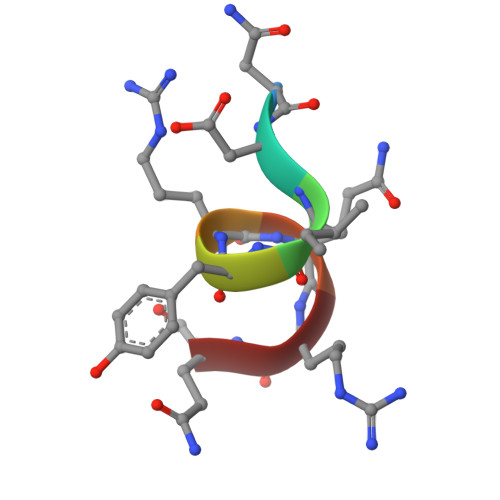
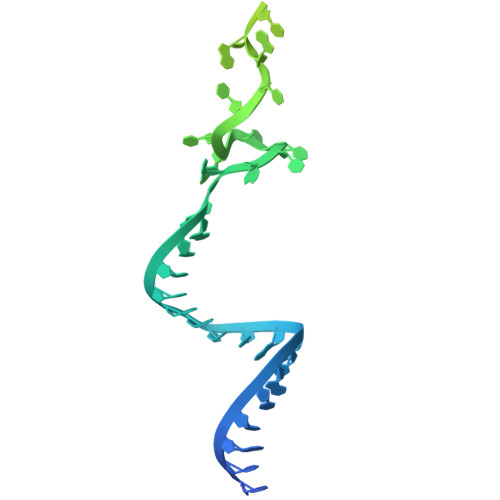
An N-terminal helix of Lsm11 stabilizes CPSF73 in U7 snRNP for histone pre-mRNA 3'-end processing.
Desotell, A., Marzluff, W.F., Dominski, Z., Tong, L.(2026) Nucleic Acids Res 54
- PubMed: 41495886
- DOI: https://doi.org/10.1093/nar/gkaf1442
- Primary Citation of Related Structures:
9N96, 9NB1, 9NGO, 9NH5, 9NH6 - PubMed Abstract:
The U7 snRNP (small nuclear ribonucleoprotein) is responsible for the 3'-end processing of replication-dependent histone messenger RNA precursors (pre-mRNAs). A helix in the Lsm11 N-terminal extension contacts the metallo-β-lactamase domain of the U7 snRNP endonuclease CPSF73. We mutated or deleted this helix and found that the mutant machineries had substantially reduced cleavage activity toward the pre-mRNA. Our cryo-electron microscopy (cryo-EM) studies indicated that the helix was important for helping to hold CPSF73 in its correct position for the cleavage reaction. We also reconstituted a wild-type U7 snRNP in complex with a methylated, noncleavable pre-mRNA. We observed that CPSF73 could achieve an open conformation independent of RNA binding to its active site. Finally, we found that a previously uninterpreted EM density for a small helix at the CPSF73-CPSF100 interface belonged to the C-terminal end of CstF77, copurified from insect cells and highly conserved among CstF77 homologs. This CstF77 binding site had a small effect on the cleavage activity of U7 snRNP. Overall, our studies have revealed the importance of the conserved helix in the Lsm11 N-terminal extension for U7 snRNP, provided structural evidence that CPSF73 can achieve an open, active conformation without RNA binding in its active site, and identified a previously unknown binding site for CstF77 in CPSF100.
- Department of Biological Sciences, Columbia University, New York, NY 10027, United States.
Organizational Affiliation: