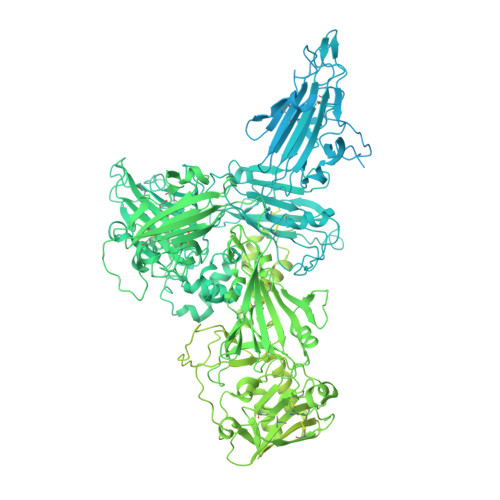
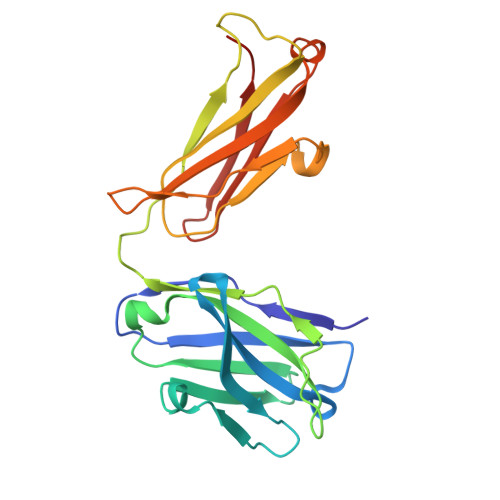
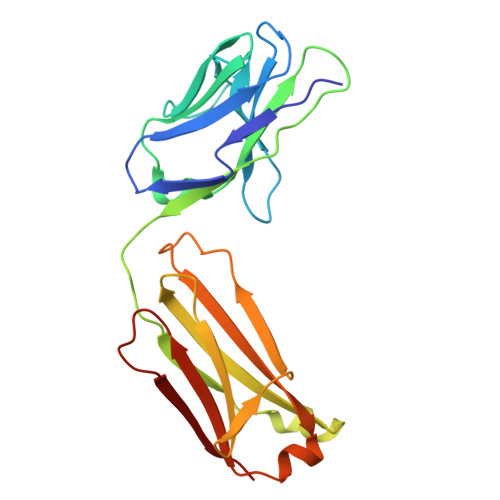
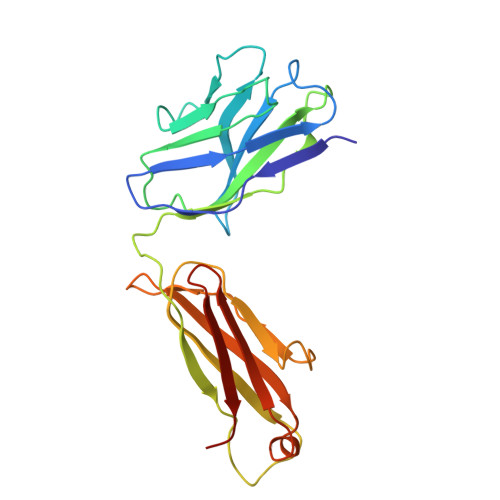
Structure of endogenous Pfs230:Pfs48/45 in complex with potent malaria transmission-blocking antibodies.
Bekkering, E.T., Yoo, R., Hailemariam, S., Heide, F., Ivanochko, D., Jackman, M., Proellochs, N.I., Stoter, R., van Gemert, G.J., Maeda, A., Yuguchi, T., Wanders, O.T., van Daalen, R.C., Inklaar, M.R., Andrade, C.M., Jansen, P.W.T.C., Vermeulen, M., Bousema, T., Takashima, E., Rubinstein, J.L., Kooij, T.W.A., Jore, M.M., Julien, J.P.(2025) bioRxiv
- PubMed: 39990443
- DOI: https://doi.org/10.1101/2025.02.14.638310
- Primary Citation of Related Structures:
9N5H, 9N5K, 9N5O - PubMed Abstract:
The Pfs230:Pfs48/45 complex forms the basis for leading malaria transmission-blocking vaccine candidates, yet little is known about its molecular assembly. Here, we used cryogenic electron microscopy to elucidate the structure of the endogenous Pfs230:Pfs48/45 complex bound to six potent transmission-blocking antibodies. Pfs230 consists of multiple domain clusters rigidified by interactions mediated through insertion domains. Membrane-anchored Pfs48/45 forms a disc-like structure and interacts with a short C-terminal peptide on Pfs230 that is critical for Pfs230 membrane-retention in vivo . Interestingly, membrane retention through this interaction is not essential for transmission to mosquitoes, suggesting that complex disruption is not a mode of action for transmission-blocking antibodies. Analyses of Pfs48/45- and Pfs230-targeted antibodies identify conserved epitopes on the Pfs230:Pfs48/45 complex and provides a structural paradigm for complement-dependent activity of Pfs230-targeting antibodies. Altogether, the antibody-bound Pfs230:Pfs48/45 structure presented improves our molecular understanding of this biological complex, informing the development of next-generation Plasmodium falciparum transmission-blocking interventions.
- Department of Medical Microbiology, Radboud University Medical Center, The Netherlands.
Organizational Affiliation: