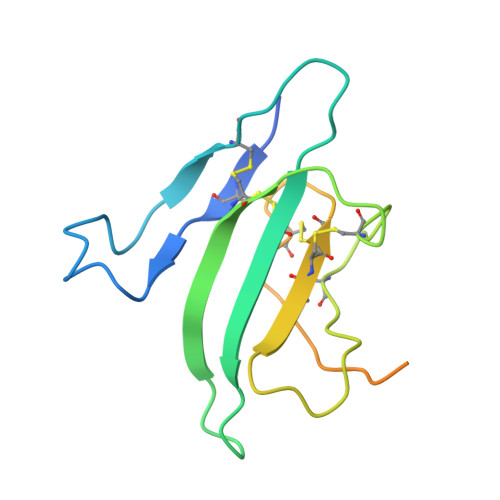
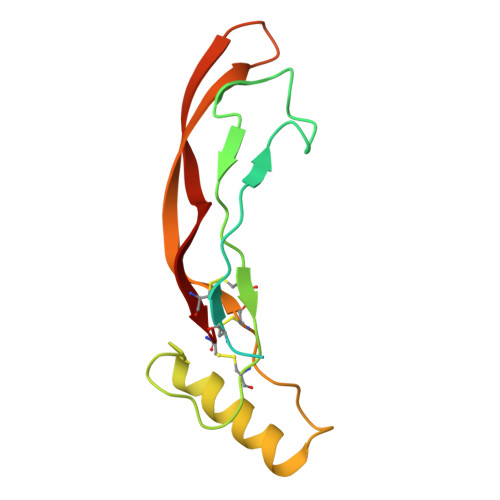
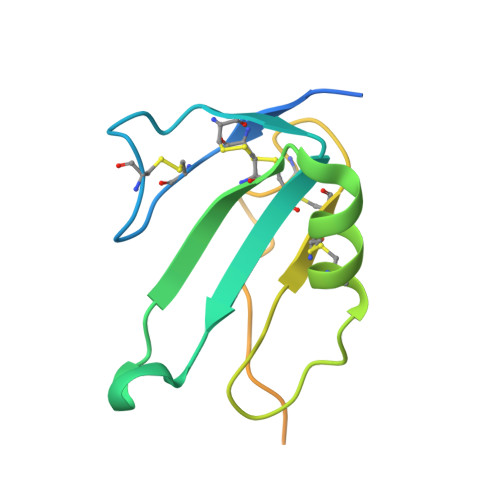
CryoEM structure of ALK2:BMP6 reveals distinct mechanism that allow ALK2 to interact with both BMP and activin ligands.
Goebel, E.J., Aykul, S., Hom, W.W., Saotome, K., Economides, A.N., Franklin, M.C., Idone, V.J.(2025) Proc Natl Acad Sci U S A 122: e2502788122-e2502788122
- PubMed: 40854140
- DOI: https://doi.org/10.1073/pnas.2502788122
- Primary Citation of Related Structures:
9MIR, 9N4K - PubMed Abstract:
Ligands in the transforming growth factor β (TGF-β) family [activins, Bone Morphogenetic Proteins (BMPs), and TGF-βs] signal by bringing together two type I and two type II receptors. Activin receptor-like kinase-2 (ALK2) is the only type I receptor among the seven TGF-β type I receptors that interacts with both activin and BMP ligands. With BMPs, ALK2 acts as a signaling receptor to activate small mothers against decapentaplegic 1 (SMAD1)/5/8 signaling. Alternatively, with activins, such as Activin A (ActA), ALK2 forms nonsignaling complexes that negatively regulate ALK2 and ActA signaling. To gain insight into how ALK2 interacts with two distinct classes of ligands, we resolved the cryoelectron microscopy structure of ALK2 in complex with the type II receptor, ActRIIB, and the ligand, BMP6, in parallel with the corresponding structure with ALK3 for direct comparison. These structures demonstrate that ALK2 and ALK3 utilize different mechanisms to interact with BMP6 at the wrist interface, with ALK2 relying on BMP6 glycosylation and ALK3 relying on a salt bridge. Modeling of ALK2:ActA reveals that binding relies on ActA's fingertip region, mirroring the interaction of ActA with its other receptor, ALK4. Our results demonstrate that ALK2 is a "hybrid" receptor that incorporates features of BMP type I receptors such as ALK3 at the wrist interface and an activin type I receptor such as ALK4 at the fingertip.
- Connective Tissue Diseases Therapeutic Focus Area, Regeneron Pharmaceuticals, Tarrytown, NY 10591.
Organizational Affiliation: