Mfd-bound E.coli RNA polymerase elongation complex + ADP-BeF3 - L1 state
Brewer, J.J., Darst, S.A., Campbell, E.A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription-repair-coupling factor | 1,166 | Escherichia coli | Mutation(s): 0 Gene Names: mfd, ACU57_04350, BGM66_002106, CTR35_002399, E4K51_18465, EIZ93_13025, FOI11_007375, FOI11_14345, FWK02_07210, GP944_10720... EC: 3.6.4 | 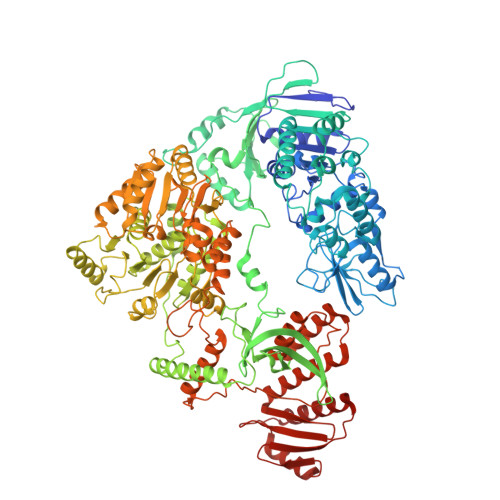 | |
UniProt | |||||
Find proteins for P30958 (Escherichia coli (strain K12)) Explore P30958 Go to UniProtKB: P30958 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P30958 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit alpha | B [auth G], C [auth H] | 234 | Escherichia coli | Mutation(s): 0 Gene Names: rpoA, FTV90_00720, FTV93_20930 EC: 2.7.7.6 | 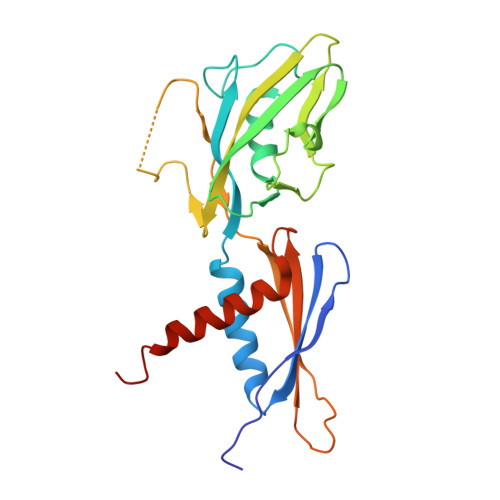 |
UniProt | |||||
Find proteins for P0A7Z4 (Escherichia coli (strain K12)) Explore P0A7Z4 Go to UniProtKB: P0A7Z4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7Z4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit beta | D [auth I] | 1,340 | Escherichia coli | Mutation(s): 0 Gene Names: rpoB EC: 2.7.7.6 | 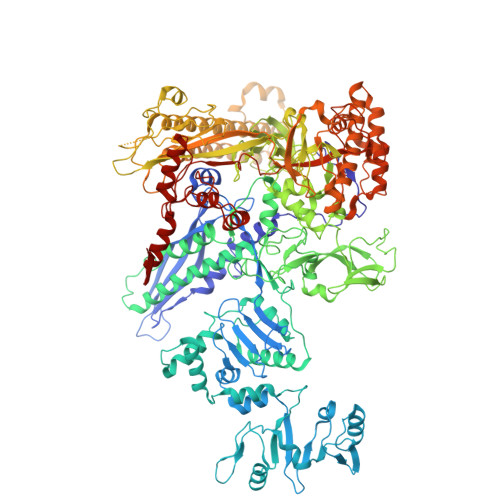 |
UniProt | |||||
Find proteins for P0A8V2 (Escherichia coli (strain K12)) Explore P0A8V2 Go to UniProtKB: P0A8V2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A8V2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit beta | E [auth J] | 1,359 | Escherichia coli | Mutation(s): 0 Gene Names: rpoC, NOI85_25215 EC: 2.7.7.6 | 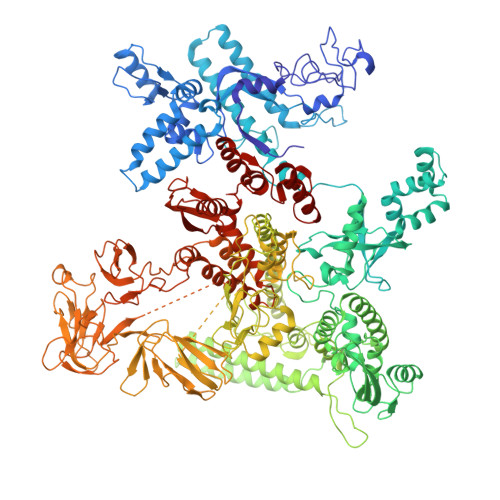 |
UniProt | |||||
Find proteins for P0A8T7 (Escherichia coli (strain K12)) Explore P0A8T7 Go to UniProtKB: P0A8T7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A8T7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit omega | F [auth K] | 91 | Escherichia coli | Mutation(s): 0 Gene Names: rpoZ, b3649, JW3624 EC: 2.7.7.6 |  |
UniProt | |||||
Find proteins for P0A800 (Escherichia coli (strain K12)) Explore P0A800 Go to UniProtKB: P0A800 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A800 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (49-MER) | G [auth N] | 64 | synthetic construct |  | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (5'-R(P*GP*GP*AP*GP*AP*GP*GP*UP*A)-3') | H [auth R] | 20 | synthetic construct |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (55-MER) | I [auth T] | 64 | synthetic construct |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP (Subject of Investigation/LOI) Query on ADP | J [auth A] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| BEF (Subject of Investigation/LOI) Query on BEF | K [auth A] | BERYLLIUM TRIFLUORIDE ION Be F3 OGIAHMCCNXDTIE-UHFFFAOYSA-K |  | ||
| ZN Query on ZN | M [auth J], N [auth J] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | L [auth J] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21.2_5419 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | -- |