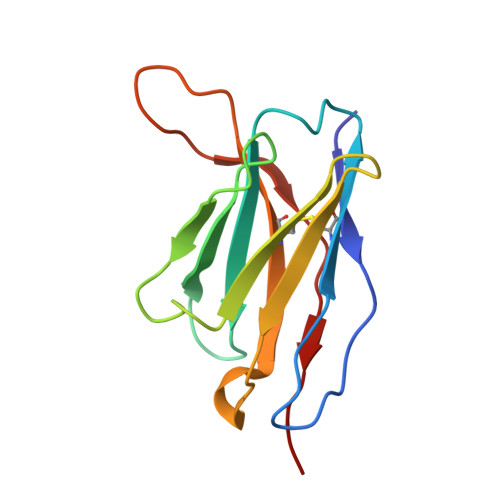
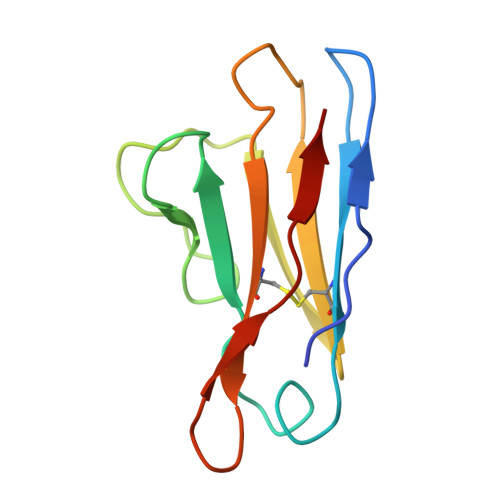
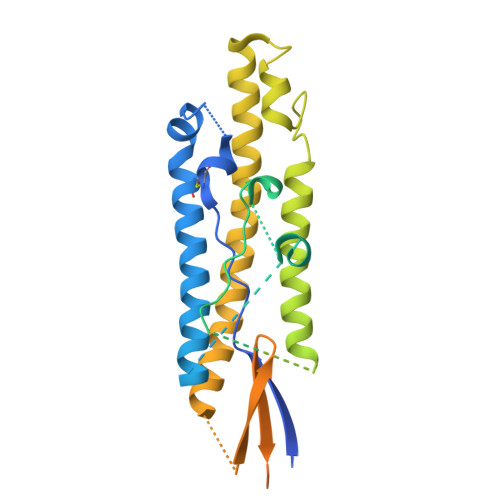
Common cold embecovirus imprinting primes broadly neutralizing antibody responses to SARS-CoV-2 S2.
Changrob, S., Yasuhara, A., Park, S., Bangaru, S., Li, L., Troxell, C.A., Halfmann, P.J., Erickson, S.A., Catanzaro, N.J., Yuan, M., Zhou, P., Huang, M., Wilbanks, G.D., McGrath, J.J.C., Singh, G., Nelson, S.A., Fu, Y., Zheng, N.Y., Carayannopoulos, S.M., Dugan, H.L., Shaw, D.G., Stamper, C.T., Madariaga, M.L.L., Krammer, F., Andrabi, R., Burton, D.R., Ward, A.B., Wilson, I.A., Kawaoka, Y., Wilson, P.C.(2025) J Exp Medicine 222
- PubMed: 41066082
- DOI: https://doi.org/10.1084/jem.20251146
- Primary Citation of Related Structures:
9MR1, 9MR2 - PubMed Abstract:
The S2 subunit of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike is highly conserved across coronavirus strains and therefore is a potential pan-coronavirus vaccine target. However, antibodies targeting this region are typically non-neutralizing. We report herein that S2-targeting antibodies from patients who recovered from SARS-CoV-2 infection bound only closely related sarbecovirus subgenus strains and, like most known S2 antibodies, none of these were neutralizing. In contrast, first-exposure, severe acutely infected COVID-19 patients predominantly induced back-boosted antibody-secreting cells imprinted against past common cold coronavirus strain OC43 that were cross-reactive to as many as five subgenera of betacoronavirus strains and gave rise to antibodies that were neutralizing and protective. The antibodies targeted two different sites: one defined by competition with stem helix antibodies, and the second to an underdescribed epitope at the apex of S2. These findings suggest that S2-targeted vaccines could strategically exploit controlled OC43 priming followed by SARS-CoV-2 boosting to enhance the breadth and quality of protective antibody responses.
- Drukier Institute for Children's Health, Weill Cornell Medicine , New York, NY, USA.
Organizational Affiliation: