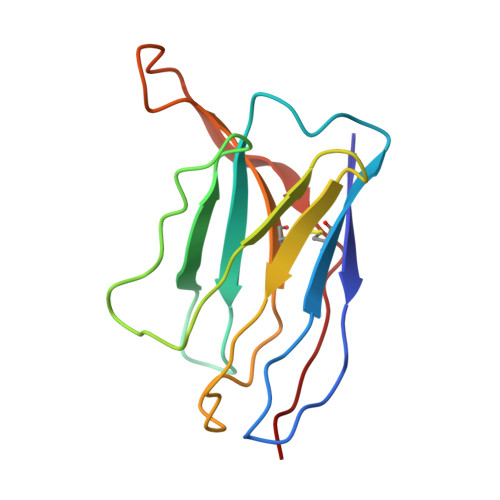
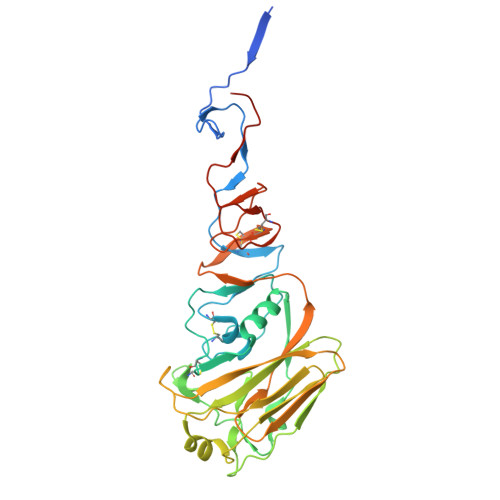
Human monoclonal antibodies that target clade 2.3.4.4b H5N1 hemagglutinin.
Alzua, G.P., Leon, A.N., Yellin, T., Bhavsar, D., Loganathan, M., Bushfield, K., Brouwer, P.J.M., Rodriguez, A.J., Jeevan, T., Webby, R., Marizzi, C., Han, J., Ward, A.B., Duty, J.A., Krammer, F.(2025) Nat Commun 17: 135-135
- PubMed: 41390501
- DOI: https://doi.org/10.1038/s41467-025-66829-y
- Primary Citation of Related Structures:
9MQ1, 9MQ2, 9MQ3 - PubMed Abstract:
The highly pathogenic avian influenza H5N1 virus clade 2.3.4.4b has been spreading globally since 2022, causing mortality and morbidity in domestic and wild birds, as well as in mammals, which underscores its potential to cause a pandemic. Here, we generate a panel of anti-hemagglutinin (HA) human monoclonal antibodies (mAbs) against the H5 protein of clade 2.3.4.4b. To develop human chimeric antibodies, H2L2 Harbor Mice®, which express human immunoglobulin germline genes, were immunized with H5 and N1 recombinant proteins from A/mallard/New York/22-008760-007- original/2022 H5N1 virus. Through hybridoma technology, sixteen fully human mAbs are generated, most of which show cross-reactivity against H5 proteins from different clade 2.3.4.4 virus variants. Fourteen out of the sixteen mAbs neutralize the virus in vitro. The mAbs with the strongest hemagglutination inhibition activity also demonstrate greater neutralizing capacity and show increased protective effects in vivo when administered prophylactically or therapeutically in a murine H5N1 challenge model. Using cryo-electron microscopy, we identify a cross-clonotype conserved motif that bound a hydrophobic groove on the head domain of H5 HA. Akin to mAbs against severe acute respiratory syndrome coronavirus 2 during the coronavirus 2019 pandemic, these mAbs could serve as treatments in case of a widespread H5N1 epidemic or pandemic.
- Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Organizational Affiliation: