Structure and function of IWS1 in transcription elongation
Syau, D., Farnung, L.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase II subunit RPB1 | 1,984 | Sus scrofa | Mutation(s): 0 | 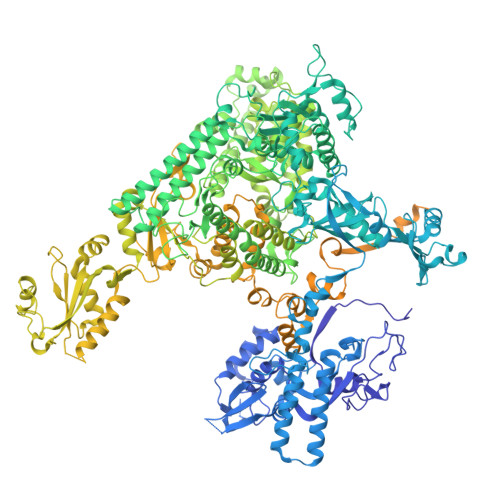 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit beta | 1,300 | Sus scrofa | Mutation(s): 0 EC: 2.7.7.6 | 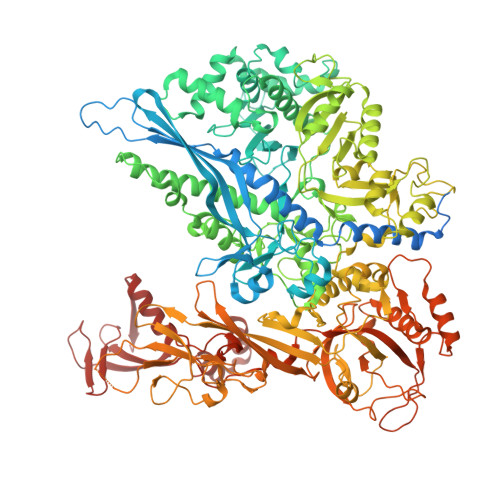 | |
UniProt | |||||
Find proteins for I3LGP4 (Sus scrofa) Explore I3LGP4 Go to UniProtKB: I3LGP4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | I3LGP4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase II subunit RPB3 | 275 | Sus scrofa | Mutation(s): 0 | 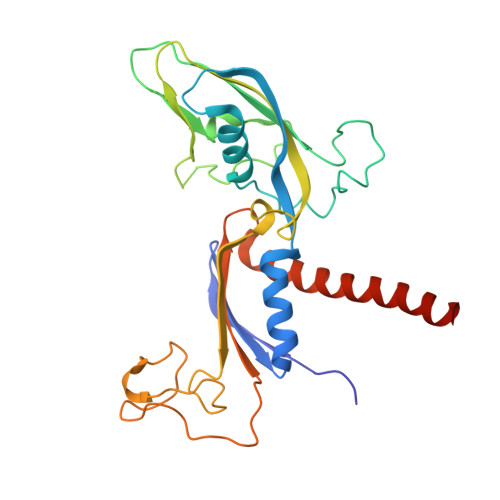 | |
UniProt | |||||
Find proteins for A0A481DF93 (Sus scrofa) Explore A0A481DF93 Go to UniProtKB: A0A481DF93 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A481DF93 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RPOL4c domain-containing protein | 184 | Sus scrofa | Mutation(s): 0 | 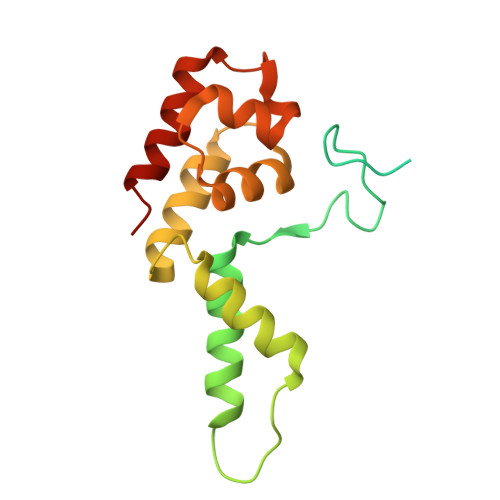 | |
UniProt | |||||
Find proteins for A0A8D1GCS3 (Sus scrofa) Explore A0A8D1GCS3 Go to UniProtKB: A0A8D1GCS3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8D1GCS3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase II subunit E | 210 | Sus scrofa | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A4X1VTX4 (Sus scrofa) Explore A0A4X1VTX4 Go to UniProtKB: A0A4X1VTX4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A4X1VTX4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerases I, II, and III subunit RPABC2 | 127 | Sus scrofa | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A4X1VEK9 (Sus scrofa) Explore A0A4X1VEK9 Go to UniProtKB: A0A4X1VEK9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A4X1VEK9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase II subunit RPB7 | 172 | Sus scrofa | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A4X1VKG7 (Sus scrofa) Explore A0A4X1VKG7 Go to UniProtKB: A0A4X1VKG7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A4X1VKG7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerases I, II, and III subunit RPABC3 | 150 | Sus scrofa | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A8D1AQR7 (Sus scrofa) Explore A0A8D1AQR7 Go to UniProtKB: A0A8D1AQR7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8D1AQR7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase II subunit RPB9 | 125 | Sus scrofa | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for P60899 (Sus scrofa) Explore P60899 Go to UniProtKB: P60899 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60899 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerases I, II, and III subunit RPABC5 | 67 | Sus scrofa | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A8W4F9W9 (Sus scrofa) Explore A0A8W4F9W9 Go to UniProtKB: A0A8W4F9W9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8W4F9W9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA polymerase II subunit J | 117 | Sus scrofa | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A8D0TE17 (Sus scrofa) Explore A0A8D0TE17 Go to UniProtKB: A0A8D0TE17 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8D0TE17 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA polymerase II subunit K | 58 | Sus scrofa | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A4X1TRS6 (Sus scrofa) Explore A0A4X1TRS6 Go to UniProtKB: A0A4X1TRS6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A4X1TRS6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription elongation factor SPT6 | 1,726 | Homo sapiens | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription elongation factor 1 homolog | 83 | Homo sapiens | Mutation(s): 0 Gene Names: ELOF1 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P60002 (Homo sapiens) Explore P60002 Go to UniProtKB: P60002 | |||||
PHAROS: P60002 GTEx: ENSG00000130165 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60002 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 17 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA polymerase-associated protein CTR9 homolog | 1,173 | Homo sapiens | Mutation(s): 0 Gene Names: CTR9, KIAA0155, SH2BP1 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q6PD62 (Homo sapiens) Explore Q6PD62 Go to UniProtKB: Q6PD62 | |||||
PHAROS: Q6PD62 GTEx: ENSG00000198730 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6PD62 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 18 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA polymerase-associated protein RTF1 homolog | 710 | Homo sapiens | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 19 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription elongation factor A protein 1 | 301 | Homo sapiens | Mutation(s): 0 Gene Names: TCEA1, GTF2S, TFIIS |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P23193 (Homo sapiens) Explore P23193 Go to UniProtKB: P23193 | |||||
PHAROS: P23193 GTEx: ENSG00000187735 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P23193 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 21 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA polymerase-associated protein LEO1 | 666 | Homo sapiens | Mutation(s): 0 Gene Names: LEO1, RDL |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8WVC0 (Homo sapiens) Explore Q8WVC0 Go to UniProtKB: Q8WVC0 | |||||
PHAROS: Q8WVC0 GTEx: ENSG00000166477 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8WVC0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 22 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA polymerase II-associated factor 1 homolog | 531 | Homo sapiens | Mutation(s): 0 Gene Names: PAF1, PD2 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8N7H5 (Homo sapiens) Explore Q8N7H5 Go to UniProtKB: Q8N7H5 | |||||
PHAROS: Q8N7H5 GTEx: ENSG00000006712 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8N7H5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 23 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| WDR61 | 305 | Homo sapiens | Mutation(s): 0 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9GZS3 (Homo sapiens) Explore Q9GZS3 Go to UniProtKB: Q9GZS3 | |||||
PHAROS: Q9GZS3 GTEx: ENSG00000140395 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9GZS3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 24 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Parafibromin | 531 | Homo sapiens | Mutation(s): 0 Gene Names: CDC73, C1orf28, HRPT2 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q6P1J9 (Homo sapiens) Explore Q6P1J9 Go to UniProtKB: Q6P1J9 | |||||
PHAROS: Q6P1J9 GTEx: ENSG00000134371 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6P1J9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 25 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription elongation factor SPT4 | 117 | Homo sapiens | Mutation(s): 0 Gene Names: SUPT4H1, SPT4H, SUPT4H |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P63272 (Homo sapiens) Explore P63272 Go to UniProtKB: P63272 | |||||
PHAROS: P63272 GTEx: ENSG00000213246 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63272 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 26 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription elongation factor SPT5 | 1,087 | Homo sapiens | Mutation(s): 0 Gene Names: SUPT5H, SPT5, SPT5H |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O00267 (Homo sapiens) Explore O00267 Go to UniProtKB: O00267 | |||||
PHAROS: O00267 GTEx: ENSG00000196235 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O00267 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 27 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein IWS1 homolog | AA [auth i] | 819 | Homo sapiens | Mutation(s): 0 Gene Names: IWS1, IWS1L |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96ST2 (Homo sapiens) Explore Q96ST2 Go to UniProtKB: Q96ST2 | |||||
PHAROS: Q96ST2 GTEx: ENSG00000163166 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96ST2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (194-MER) | 206 | synthetic construct |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (5'-R(P*UP*UP*UP*GP*GP*UP*GP*UP*GP*UP*CP*UP*GP*GP*GP*UP*GP*GP*UP*G)-3') | 39 | synthetic construct |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 20 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (194-MER) | 215 | synthetic construct |  | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN Query on ZN | BA [auth A] CA [auth A] EA [auth B] FA [auth C] GA [auth I] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | DA [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 3.2.0 |
| RECONSTRUCTION | Warp | |
| MODEL REFINEMENT | PHENIX | 1.20.1-4487 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Richard and Susan Smith Family Foundation | United States | -- |
| Damon Runyon Cancer Research Foundation | United States | -- |
| National Institutes of Health/National Institute of Environmental Health Sciences (NIH/NIEHS) | United States | DP2-ES036404 |