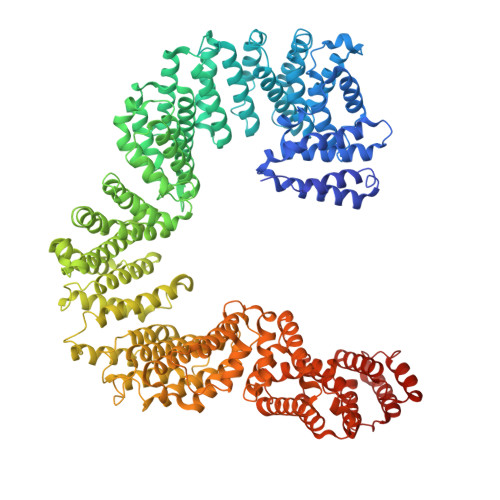
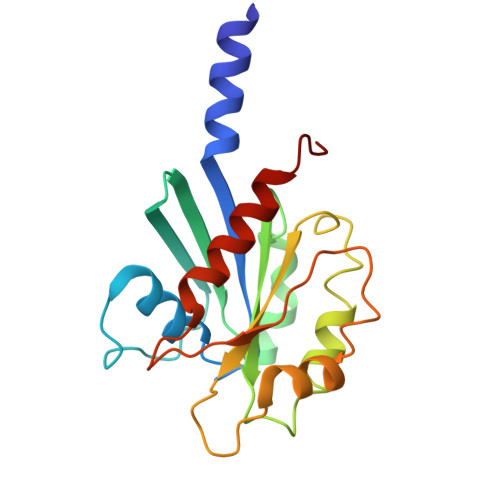
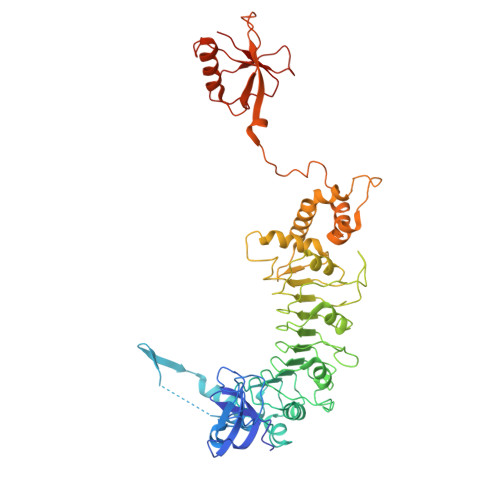
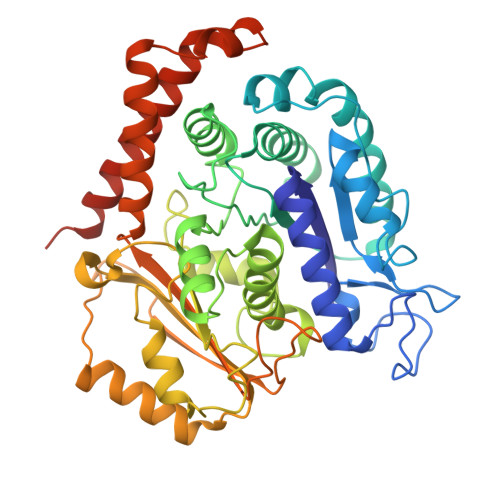
Structural dissection of alpha beta-tubulin heterodimer assembly and disassembly by human tubulin-specific chaperones.
Seong, Y., Kim, H., Byun, K., Park, Y.W., Roh, S.H.(2025) Science 390: eady2708-eady2708
- PubMed: 41166473
- DOI: https://doi.org/10.1126/science.ady2708
- Primary Citation of Related Structures:
9M1I, 9M1J, 9M1K, 9M1L, 9M1M, 9M1N - PubMed Abstract:
Microtubule assembly requires a set of chaperones known as tubulin-binding cofactors (TBCs). We used cryo-electron microscopy to visualize how human TBCD, TBCE, TBCC, and guanosine triphosphatase (GTPase) Arl2 mediate αβ-tubulin assembly and disassembly. We captured multiple conformational states, revealing how TBCs orchestrate tubulin heterodimer biogenesis. TBCD stabilizes monomeric β-tubulin and scaffolds the other cofactors. Guanosine triphosphate (GTP) binding to Arl2 induces conformational changes that toggle the complex between assembly and disassembly. TBCD and TBCE guide α- and β-tubulin into a partially assembled interface, and TBCC, acting as a molecular clamp, completes the heterodimer. TBCD also functions as a GTPase activating protein for β-tubulin. β-tubulin GTP hydrolysis is coupled to Arl2's GTPase activity, establishing a checkpoint that ensures that only fully matured heterodimers proceed. These findings provide a structural framework for tubulin heterodimer biogenesis and recycling, supporting cytoskeletal proteostasis.
- School of Biological Sciences, Seoul National University, Seoul, Republic of Korea.
Organizational Affiliation: