Structural basis for sequence-specific DNA recognition by a group IId WRKY transcription factor GhWRKY17 in cotton.
Xiao, Q., Wang, Y., Shang, X., Chen, Y., Zhang, M., Zhou, Y., Huang, X., Qin, S., Min, J., Xu, G., Liu, Y.(2026) Biochem J 483
- PubMed: 41569870
- DOI: https://doi.org/10.1042/BCJ20250191
- Primary Citation of Related Structures:
9M0K - PubMed Abstract:
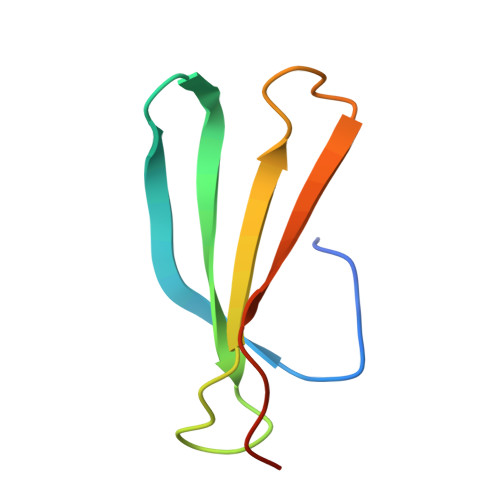
WRKY transcription factors, a plant-specific family of transcriptional regulators, are classified into four groups (I-IV) and play pivotal roles in plant defense, development, and stress responses. These proteins are characterized by conserved WRKY domains that preferentially bind to the W-box cis-element C/TTGACC/T in target gene promoters. In Gossypium hirsutum (Gh; upland cotton), the group IId member GhWRKY17 regulates cotton fiber development by activating downstream target genes such as GhHOX3 through promoter W-box binding. However, the structural basis for its DNA recognition specificity remains elusive. Here, we present the 1.8 Å resolution crystal structure of the GhWRKY17 WRKY domain in complex with the GhHOX3 promoter DNA-the first structural characterization of a group IId WRKY protein. Structural analysis reveals that it consists of four antiparallel β-strands, with the β2-strand (harboring the conserved 249WRKYGQK255 motif) and β3-strand co-operatively engaging the DNA major groove. Key residues (R250, K251, Y252, Q254, K255, R264, Y266, Y267) form an intricate hydrogen-bonding network essential for recognizing the extended G/TTTGACC motif. Comparative structural analyses with group I/IIa/III WRKY-DNA complexes reveal that GhWRKY17's dual-strand engagement and extensively hydrogen bond-mediated specific interaction represent novel mechanistic features distinguishing group IId members from other WRKY subgroups, emphasizing the necessity for subgroup-specific investigations. These findings not only establish a structural paradigm for group IId WRKY function but also provide molecular insights for engineering cotton fiber traits through transcriptional regulation.
- Jiangsu Key Laboratory of Drug Discovery and Translational Research for Brain Diseases, College of Pharmaceutical Sciences, Soochow University, Suzhou, Jiangsu, 215123, China.
Organizational Affiliation: