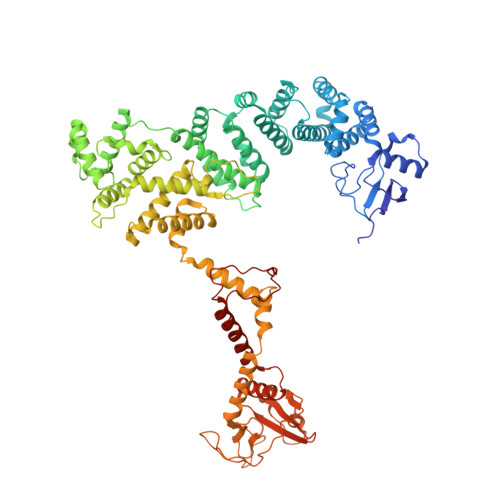
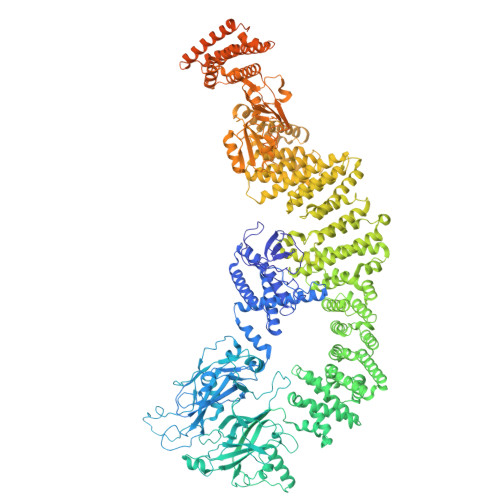
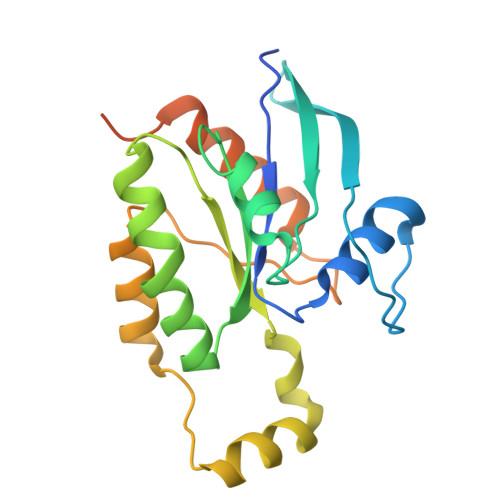
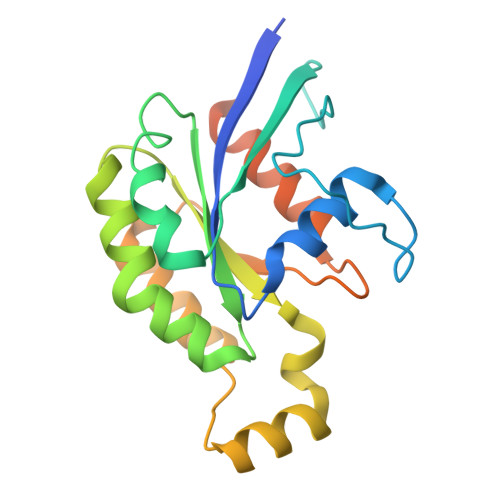
Conformational alteration of DOCK5•ELMO1 signalosome on lipid membrane.
Shinoda, T., Katsura, K., Ishizuka-Katsura, Y., Hanada, K., Yonemochi, M., Miyamoto, Y., Kukimoto-Niino, M., Yamauchi, J., Shirouzu, M.(2025) Commun Biol 8: 1523-1523
- PubMed: 41233496
- DOI: https://doi.org/10.1038/s42003-025-09113-5
- Primary Citation of Related Structures:
9LX0, 9LXH - PubMed Abstract:
The DOCK protein family activates Rho small GTPases through guanine nucleotide exchange factor (GEF) activity. DOCK is thought to exert its GEF activity at the plasma membrane. However, the mechanism by which DOCK activity on the plasma membrane is regulated remains unclear. Herein, we present a new conformation in which DOCK5, ELMO1, RhoG, and Rac1 are aligned on a plane and symmetrically flattened, as revealed by cryo-EM using a lipid membrane-coated grid. The major conformational change leading to this structure results from rotation of each DOCK5•ELMO1 hinge site through interactions with the membrane. Biochemical and cellular experiments indicate that conformational changes driven by acidic lipids are important for regulating the GEF activity of the DOCK5•ELMO1 complex on the plasma membrane and are essential for its downstream signalling. This approach also enables the analysis of large lipid-associated complexes, such as signalosomes, and will aid studies of membrane-dependent signalling assemblies.
- Laboratory for Functional and Structural Biology, RIKEN Center for Integrative Medical Sciences, Yokohama, Kanagawa, Japan.
Organizational Affiliation: