LGP2:MDA5:dsRNA filament
Kurihara, N., Isayama, Y., Nureki, O., Kato, K.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP-dependent RNA helicase DHX58 | C [auth A] | 678 | Homo sapiens | Mutation(s): 1 Gene Names: DHX58, D11LGP2E, LGP2 EC: 3.6.4.13 | 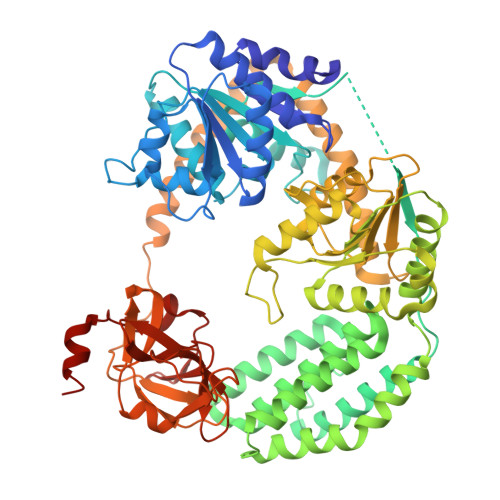 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96C10 (Homo sapiens) Explore Q96C10 Go to UniProtKB: Q96C10 | |||||
PHAROS: Q96C10 GTEx: ENSG00000108771 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96C10 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Interferon-induced helicase C domain-containing protein 1 | D [auth B], E [auth C] | 739 | Homo sapiens | Mutation(s): 3 Gene Names: IFIH1, MDA5, RH116 EC: 3.6.4.13 | 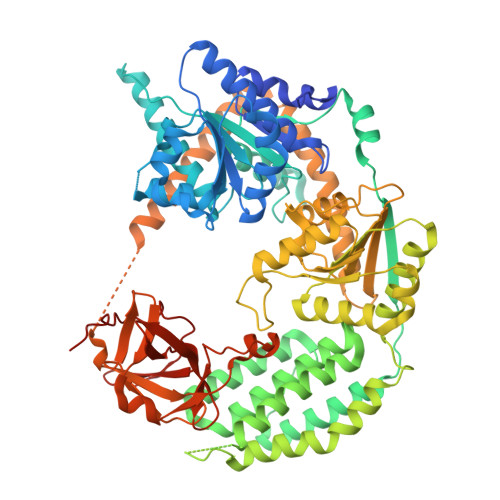 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9BYX4 (Homo sapiens) Explore Q9BYX4 Go to UniProtKB: Q9BYX4 | |||||
PHAROS: Q9BYX4 GTEx: ENSG00000115267 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9BYX4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (45-MER) | A [auth Y] | 45 | uncultured archaeon |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (45-MER) | B [auth X] | 45 | uncultured archaeon |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP Query on ADP | F [auth A], I [auth B], L [auth C] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| ZN Query on ZN | G [auth A], J [auth B], M [auth C] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | H [auth A], K [auth B], N [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21.1_5286 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | 23H04768 |