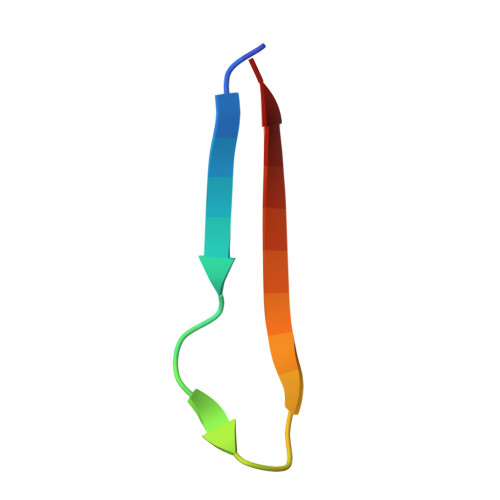
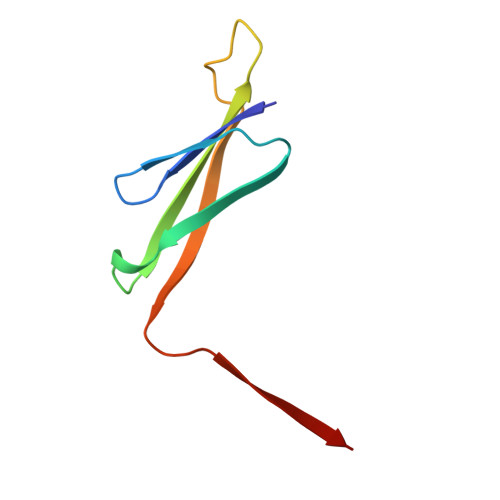
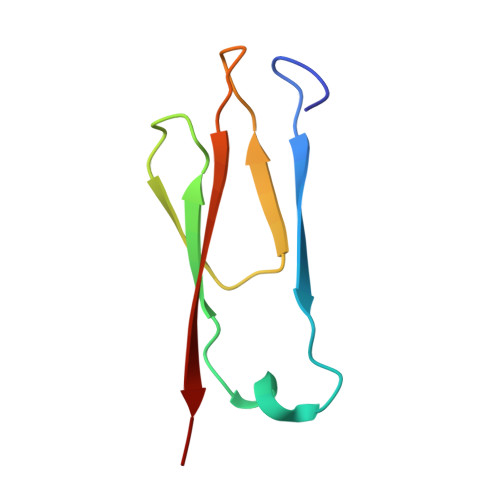
Atomic structure and in situ visualization of native PMEL lamellae in melanosomes.
Ma, B., Yao, Y., Dong, H., Yang, L., Li, D., Zhao, Q., Sun, B., Chen, Y., Liu, C., Li, D.(2025) Nat Commun 16: 10300-10300
- PubMed: 41271718
- DOI: https://doi.org/10.1038/s41467-025-65221-0
- Primary Citation of Related Structures:
9LIP - PubMed Abstract:
Melanin synthesis within melanosomes critically depends on the fibrillar architecture formed by the pigment cell-specific protein PMEL. Although PMEL fibrils have historically been classified as functional amyloids, their native supramolecular organization in situ and detailed molecular architecture have remained unresolved. In this study, we combine in situ cryo-electron tomography (cryo-ET) and cryo-electron microscopy (cryo-EM) to elucidate the native structural organization of PMEL fibrils within human melanosomes from both patient melanoma tissues and melanocyte cell lines. We demonstrate that PMEL does not form conventional isolated amyloid fibrils, but rather assembles into highly organized lamellar sheets consisting of laterally aligned fibrils interconnected by flexible linker regions. Cryo-EM structures reveal a distinctive butterfly-shaped fibril unit composed of multiple structured domains, including both the proteolytically cleaved Mα and Mβ fragments of PMEL, which assemble into a amyloid-like β-sheet arrangement. Notably, we identify intrinsically disordered regions critical for lamellar assembly and curvature and verify key glycosylation modifications in the structure. This architecture distinguishes PMEL fibrils from canonical amyloids and elucidates the molecular basis underlying melanosome integrity and pigmentation. Moreover, our work provides molecular insights relevant for pigmentation disorders and PMEL-associated diseases, including melanoma.
- Interdisciplinary Research Center on Biology and Chemistry, State Key Laboratory of Chemical Biology, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: