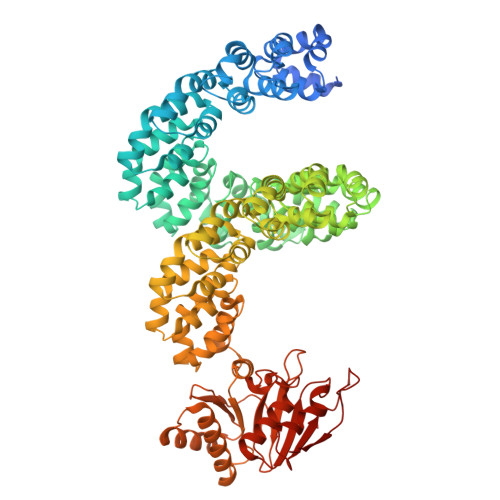
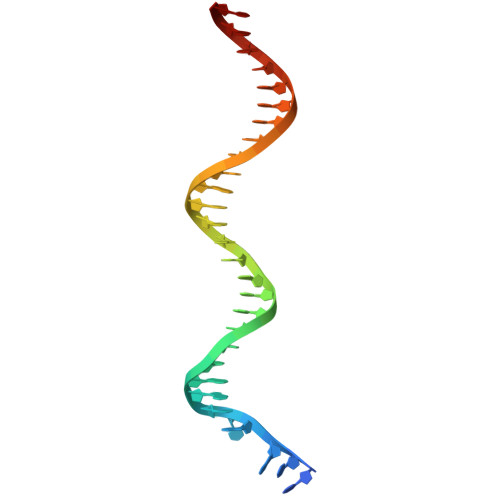
Computational design of a high-precision mitochondrial DNA cytosine base editor.
Mi, L., Li, Y.X., Lv, X., Wan, Z.L., Liu, X., Zhang, K., Li, H., Yao, Y., Zhang, L., Xu, Z., Zhuang, X., Ji, K., Jiang, M., Wang, Y., Lu, P.(2025) Nat Struct Mol Biol 32: 2575-2586
- PubMed: 41249818
- DOI: https://doi.org/10.1038/s41594-025-01714-2
- Primary Citation of Related Structures:
9LCX, 9LCY, 9LCZ, 9LD0, 9LD1 - PubMed Abstract:
Bystander editing remains a major limitation of current base editors, hindering their precision and therapeutic potential. Here, we present a de novo protein design strategy that creates a structurally rigid interface between a DNA-binding TALE domain and a cytosine deaminase, forming a unified editing module termed TALE-oriented deaminase (TOD). Cryo-EM analysis of TOD-DNA complexes confirms that this precise spatial architecture tightly restricts the deaminase activity window, thereby minimizing unwanted deamination. To further enhance editing specificity, we develop a split version, termed DdCBE-TOD, which virtually eliminates off-target editing. As a proof of concept, we apply DdCBE-TOD to generate a mitochondrial disease mouse model and to correct a pathogenic mutation associated with MERRF syndrome in patient-derived cells, achieving single-nucleotide precision. This work introduces a generalizable and computationally guided approach for ultra-precise base editing, offering a promising platform for both mechanistic studies and therapeutic correction of single-nucleotide mutations.
- State Key Laboratory of Gene Expression, Research Center for Industries of the Future, School of Life Sciences, Westlake University, Hangzhou, China.
Organizational Affiliation: