Structural basis of menaquinone reduction by succinate dehydrogenase from early photosynthetic bacterium Chloroflexus aurantiacus
Zhang, X., Wu, J.Y., Xu, X.L.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Succinate dehydrogenase or fumarate reductase, flavoprotein subunit | 657 | Chloroflexus aurantiacus J-10-fl | Mutation(s): 0 EC: 1.3.99.1 (PDB Primary Data), 1.3.5.1 (UniProt) | 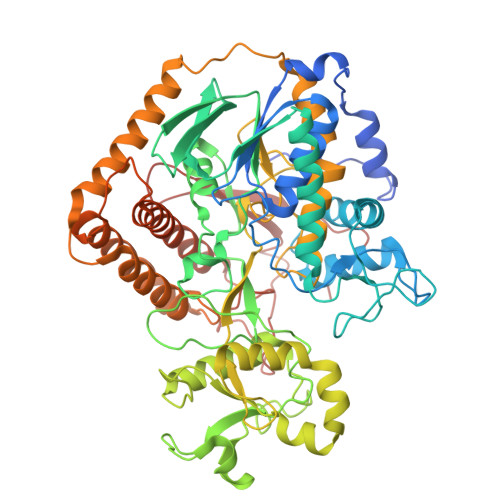 | |
UniProt | |||||
Find proteins for A9WDT7 (Chloroflexus aurantiacus (strain ATCC 29366 / DSM 635 / J-10-fl)) Explore A9WDT7 Go to UniProtKB: A9WDT7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A9WDT7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 4Fe-4S ferredoxin iron-sulfur binding domain protein | 260 | Chloroflexus aurantiacus J-10-fl | Mutation(s): 0 | 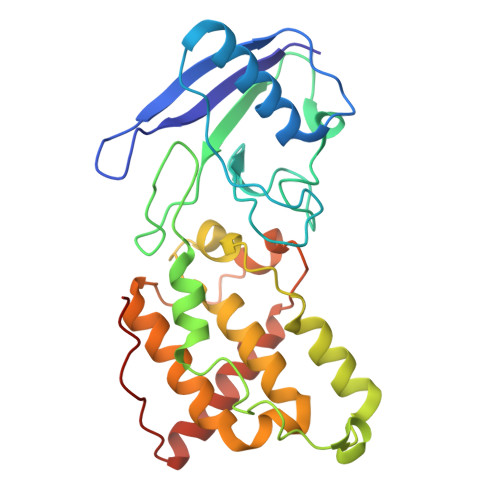 | |
UniProt | |||||
Find proteins for A9WDT6 (Chloroflexus aurantiacus (strain ATCC 29366 / DSM 635 / J-10-fl)) Explore A9WDT6 Go to UniProtKB: A9WDT6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A9WDT6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Succinate dehydrogenase (Or fumarate reductase) cytochrome b subunit, b558 family | 239 | Chloroflexus aurantiacus J-10-fl | Mutation(s): 0 | 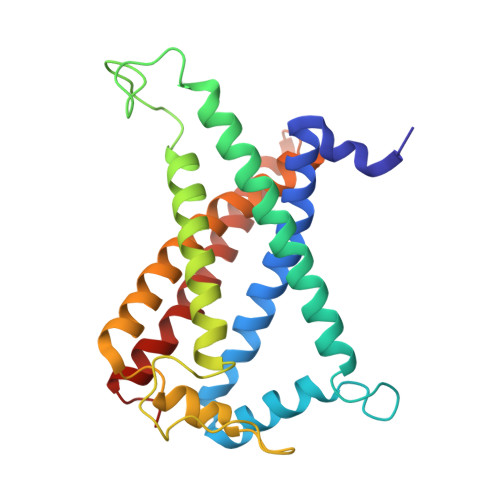 | |
UniProt | |||||
Find proteins for A9WDT8 (Chloroflexus aurantiacus (strain ATCC 29366 / DSM 635 / J-10-fl)) Explore A9WDT8 Go to UniProtKB: A9WDT8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A9WDT8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD (Subject of Investigation/LOI) Query on FAD | BA [auth G], J [auth A], S [auth D] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| MQ7 (Subject of Investigation/LOI) Query on MQ7 | AA [auth F] IA [auth I] JA [auth I] Q [auth C] R [auth C] | MENAQUINONE-7 C46 H64 O2 RAKQPZMEYJZGPI-LJWNYQGCSA-N |  | ||
| HEM (Subject of Investigation/LOI) Query on HEM | FA [auth I] GA [auth I] N [auth C] O [auth C] W [auth F] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| LMT (Subject of Investigation/LOI) Query on LMT | HA [auth I], P [auth C], Y [auth F] | DODECYL-BETA-D-MALTOSIDE C24 H46 O11 NLEBIOOXCVAHBD-QKMCSOCLSA-N |  | ||
| SF4 (Subject of Investigation/LOI) Query on SF4 | CA [auth H], K [auth B], T [auth E] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| F3S (Subject of Investigation/LOI) Query on F3S | EA [auth H], M [auth B], V [auth E] | FE3-S4 CLUSTER Fe3 S4 FCXHZBQOKRZXKS-UHFFFAOYSA-N |  | ||
| FES (Subject of Investigation/LOI) Query on FES | DA [auth H], L [auth B], U [auth E] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20.1_4487: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32471270 |
| National Natural Science Foundation of China (NSFC) | China | 32171227 |
| National Natural Science Foundation of China (NSFC) | China | 31870740 |