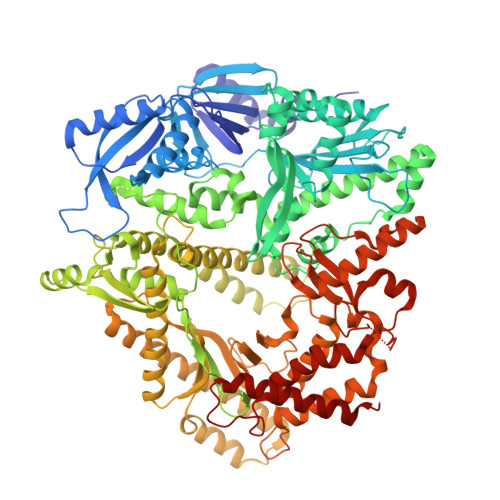
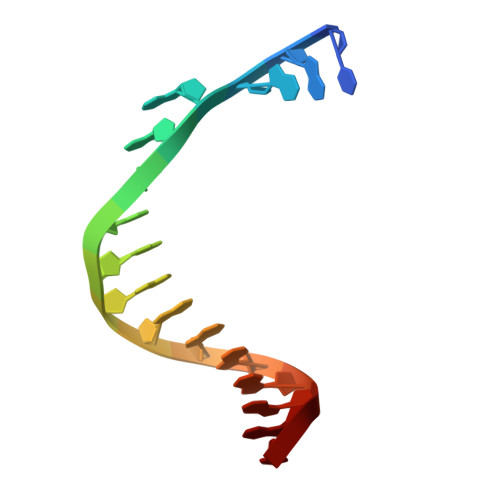
Structural basis for DNA replication by the African swine fever virus polymerase.
Hu, Z., SunKang, Y., Liu, Z., Huang, L., Qi, W., Yan, R.(2025) iScience 28: 114155-114155
- PubMed: 41438038
- DOI: https://doi.org/10.1016/j.isci.2025.114155
- Primary Citation of Related Structures:
9L8T, 9L8U - PubMed Abstract:
African swine fever virus (ASFV) devastates swine herds and lacks widely available antivirals. We show that the ASFV DNA polymerase beta (Pol β; gene G1211R ) localizes to viral replication factories and is required for viral replication. Cryo-electron microscopy structures of Pol β in apo and double-stranded DNA (dsDNA)-bound states reveal pronounced conformational flexibility that enables transitions between functional states. Binding to dsDNA promotes higher-order oligomerization that is essential for catalytic activity, as demonstrated by structure-guided mutagenesis and biochemical assays. These findings define the structural basis of ASFV genome synthesis, highlight the functional significance of Pol β during viral replication, and provide a framework for polymerase-targeted antiviral discovery.
- SUSTech Homeostatic Medicine Institute, School of Medicine, Southern University of Science and Technology, Shenzhen, Guangdong Province, China.
Organizational Affiliation: