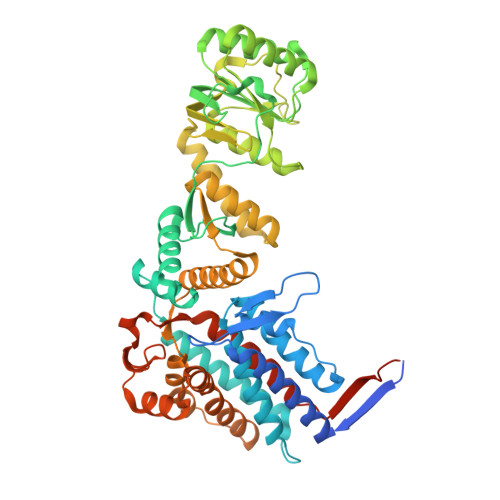
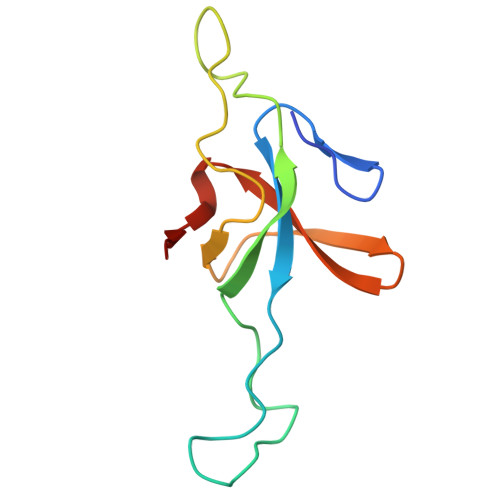
In situ characterization of mitochondrial Hsp60-Hsp10 chaperone complex under folding stress.
Jung, M., Kim, M., Ham, S.J., Chung, J., Roh, S.H.(2025) Sci Adv 11: eadw6064-eadw6064
- PubMed: 41124257
- DOI: https://doi.org/10.1126/sciadv.adw6064
- Primary Citation of Related Structures:
9L8P - PubMed Abstract:
Mitochondrial proteostasis is critical for maintaining mitochondrial function, and its disruption induces mitochondrial unfolded protein response, which up-regulates chaperones to alleviate protein-folding stress. However, how these chaperones mitigate protein-folding stress remains unclear. Here, using correlated cryo-electron tomography, we show that folding stress triggers marked mitochondrial morphological changes, including the accumulation of amorphous protein aggregates and increased abundance and spatial clustering of the mitochondrial heat shock protein 60-heat shock protein 10 (mtHsp60-Hsp10) complex. Subtomogram analysis revealed the in situ architecture and conformational heterogeneity of mtHsp60-Hsp10 under stress, which retains its canonical double-ring structure while adopting distinct football, half-football, and bullet-like states. Notably, the mtHsp60-Hsp10 complex encapsulates unstructured substrates through conserved hydrophobic interactions. We further demonstrate that knockdown of the mtHsp60-Hsp10 complex exacerbates folding stress, as evidenced by elevated cellular stress responses and activation of mitophagy. Our study defines the in situ structural properties of the mtHsp60-Hsp10 complex and provides mechanistic insight into how it safeguards mitochondrial proteostasis under folding stress.
- School of Biological Sciences, Seoul National University, Seoul 08826, Republic of Korea.
Organizational Affiliation: