Hexamer Msp1 from S.cerevisiaec(with a catalytic dead mutation) in complex with an unknown peptide substrate
Chengdong, H., Simin, W., Xuan, C.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Outer mitochondrial transmembrane helix translocase | 362 | Saccharomyces cerevisiae S288C | Mutation(s): 1 Gene Names: MSP1, YTA4, YGR028W EC: 7.4.2 | 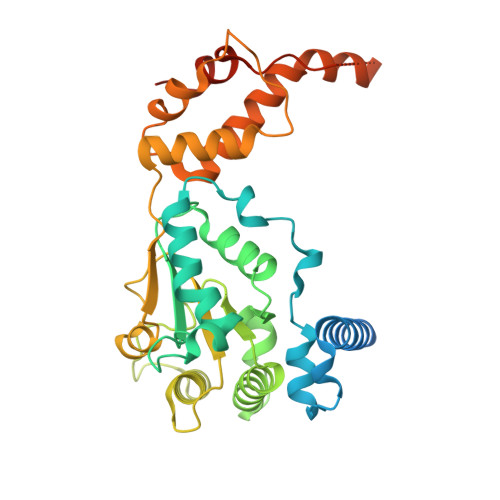 | |
UniProt | |||||
Find proteins for P28737 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P28737 Go to UniProtKB: P28737 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P28737 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| An unknown peptide substrate | 12 | Escherichia coli BL21(DE3) | Mutation(s): 0 |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP (Subject of Investigation/LOI) Query on ATP | H [auth B] J [auth F] L [auth E] N [auth D] P [auth C] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | I [auth B] K [auth F] M [auth E] O [auth D] Q [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.19_4092: |
| RECONSTRUCTION | cryoSPARC | 3.3.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |