Cryo-EM structure of human lipid phosphate phosphatase 1
Yang, M., Qian, H.W.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Phospholipid phosphatase 1 | 284 | Homo sapiens | Mutation(s): 0 Gene Names: PLPP1, LPP1, PPAP2A EC: 3.1.3 (PDB Primary Data), 3.1.3.106 (PDB Primary Data), 3.1.3.4 (PDB Primary Data), 3.6.1.75 (PDB Primary Data) | 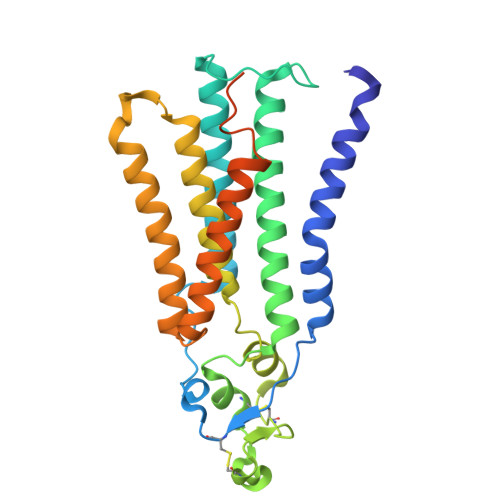 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O14494 (Homo sapiens) Explore O14494 Go to UniProtKB: O14494 | |||||
PHAROS: O14494 GTEx: ENSG00000067113 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O14494 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: O14494-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PT5 (Subject of Investigation/LOI) Query on PT5 | H [auth A], L [auth B], P [auth C], T [auth D] | [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho
ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate C47 H85 O19 P3 CNWINRVXAYPOMW-HJBQCNPJSA-N |  | ||
| LBN (Subject of Investigation/LOI) Query on LBN | G [auth A], K [auth B], O [auth C], S [auth D] | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine C42 H82 N O8 P WTJKGGKOPKCXLL-VYOBOKEXSA-N |  | ||
| LPP (Subject of Investigation/LOI) Query on LPP | E [auth A], I [auth B], M [auth C], Q [auth D] | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE C35 H69 O8 P PORPENFLTBBHSG-MGBGTMOVSA-N |  | ||
| NAG (Subject of Investigation/LOI) Query on NAG | F [auth A], J [auth B], N [auth C], R [auth D] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Chinese Academy of Sciences | China | -- |