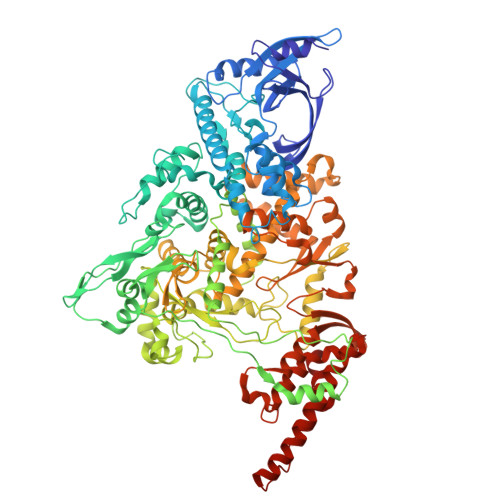
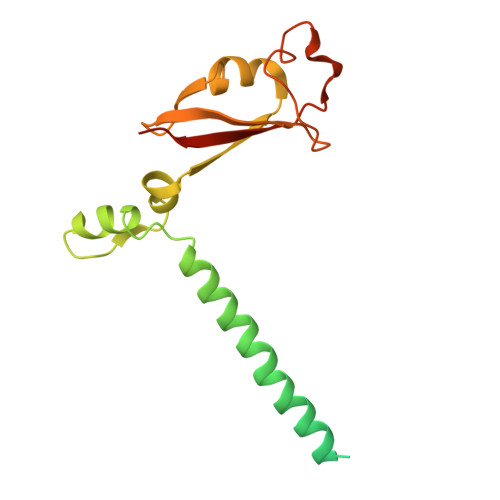
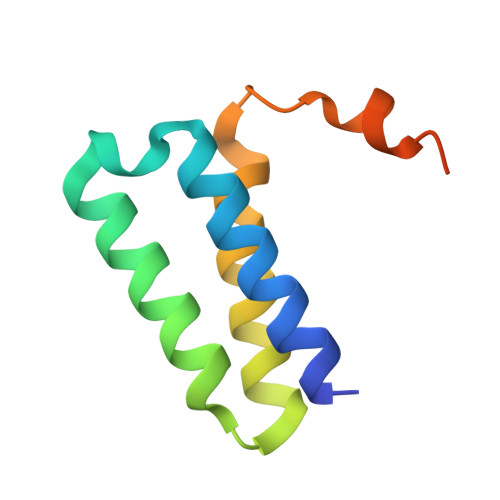
Discovery of a 2'-a-Fluoro-2'-b-C-(fluoromethyl) Purine Nucleotide Prodrug as a Potential Oral Anti-SARS-CoV-2 Agent
Liang, L., Meng, Y., Chang, X., Li, E., Huang, Y., Yan, L., Lou, Z., Peng, Y., Zhu, B., Yu, W., Chang, J.(2025) J Med Chem 68: 1994-2007
- PubMed: 39804580
- DOI: https://doi.org/10.1021/acs.jmedchem.4c02769
- Primary Citation of Related Structures:
9L09 - PubMed Abstract:
A novel 2'-α-fluoro-2'-β- C -(fluoromethyl) purine nucleoside phosphoramidate prodrug 15 has been designed and synthesized to treat SARS-CoV-2 infection. The SARS-CoV-2 central replication transcription complex (C-RTC, nsp12-nsp7-nsp8 2 ) catalyzed in vitro RNA synthesis was effectively inhibited by the corresponding bioactive nucleoside triphosphate ( 13-TP ). The cryo-electron microscopy structure of the C-RTC: 13-TP complex was also determined. Compound 15 exhibited potent in vitro antiviral activity against the SARS-CoV-2 20SF107 strain (EC 50 = 0.56 ± 0.06 μM) and the Omicron BA.5 variant (EC 50 = 0.96 ± 0.23 μM) with low cytotoxicity. Furthermore, it was well tolerated in rats at doses of up to 2000 mg/kg, and a single oral dose of this prodrug at 40 mg/kg led to high levels of 13-TP in the target organ lungs of rats with a long half-life. These findings support the further development of compound 15 as an orally available antiviral agent for the treatment of SARS-CoV-2 infection.
- State Key Laboratory of Antiviral Drugs, Pingyuan Laboratory, NMPA Key Laboratory for Research and Evaluation of Innovative Drug, School of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang, Henan 453007, China.
Organizational Affiliation: