Cryo-EM structure of the human TRPC1/C5 heteromer
Chen, Y.X., Cheng, X.Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Maltose/maltodextrin-binding periplasmic protein,Short transient receptor potential channel 5 | 1,203 | Escherichia coli K-12, Mus musculus This entity is chimeric | Mutation(s): 0 Gene Names: malE, b4034, JW3994, Trpc5, Trp5, Trrp5 | 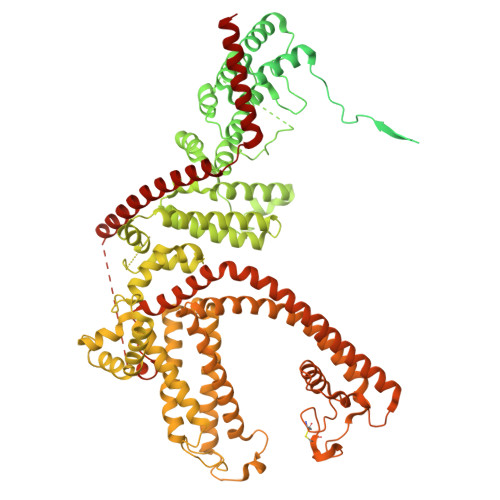 | |
UniProt | |||||
Find proteins for Q9QX29 (Mus musculus) Explore Q9QX29 Go to UniProtKB: Q9QX29 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9QX29 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Maltose/maltodextrin-binding periplasmic protein,Short transient receptor potential channel 1,Extended tegument protein pp150 | 1,504 | Escherichia coli K-12, Homo sapiens, Human betaherpesvirus 5 This entity is chimeric | Mutation(s): 0 Gene Names: malE, b4034, JW3994, TRPC1, TRP1, UL32 | 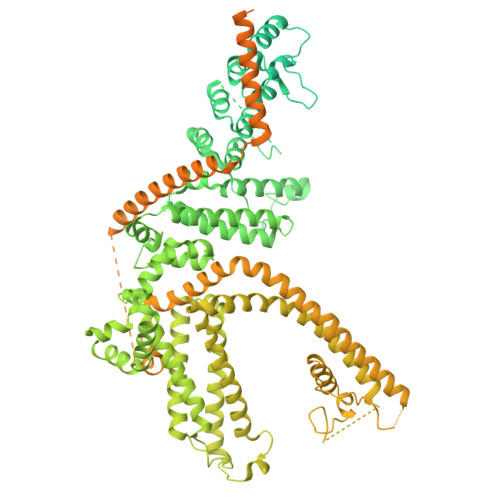 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for A0A076JQ90 (Human cytomegalovirus) Explore A0A076JQ90 Go to UniProtKB: A0A076JQ90 | |||||
Find proteins for P48995 (Homo sapiens) Explore P48995 Go to UniProtKB: P48995 | |||||
PHAROS: P48995 GTEx: ENSG00000144935 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | A0A076JQ90P48995 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| YZY (Subject of Investigation/LOI) Query on YZY | G [auth A], H [auth B], K [auth C], L [auth C] | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate C37 H70 O5 DOZKMFVMCATMEH-OZKTZCCCSA-N |  | ||
| Y01 (Subject of Investigation/LOI) Query on Y01 | F [auth A], J [auth C], N [auth D] | CHOLESTEROL HEMISUCCINATE C31 H50 O4 WLNARFZDISHUGS-MIXBDBMTSA-N |  | ||
| CA (Subject of Investigation/LOI) Query on CA | E [auth A], I [auth C], M [auth D] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |