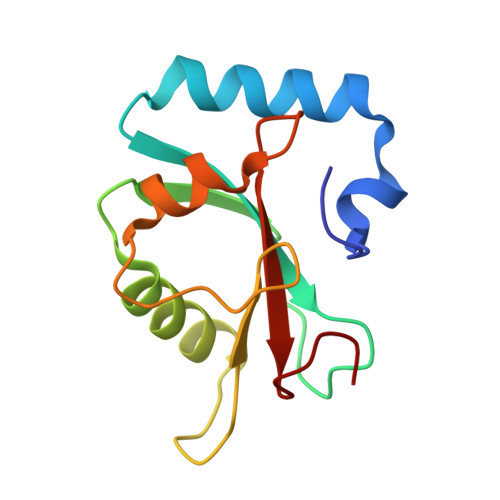
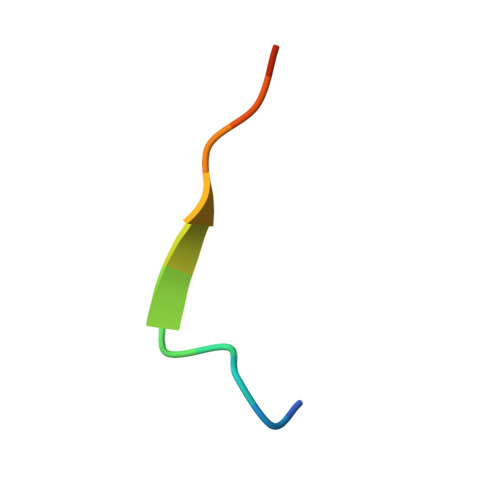
Molecular bases of the interactions of ATG16L1 with FIP200 and ATG8 family proteins.
Gong, X., Zhou, Y., Wang, Y., Tang, Y., Liu, H., Zhou, X., Zhang, Y., Guo, H., Guo, Z., Pan, L.(2025) Nat Commun 16: 9035-9035
- PubMed: 41073433
- DOI: https://doi.org/10.1038/s41467-025-64097-4
- Primary Citation of Related Structures:
9J54, 9JF2 - PubMed Abstract:
Macroautophagy maintains cellular and organismal homeostasis, and entails de novo synthesis of double-membrane autophagosome. The effective formation of autophagosome requires the recruitment of the ATG12~ATG5-ATG16L1 complex to the pre-autophagosomal structure by relevant ATG16L1-binding autophagic factors including FIP200. However, the molecular mechanism governing the specific interaction of ATG16L1 with FIP200 remains elusive. Here, we uncover that ATG16L1 contains a FIP200-interacting region (FIR), which not only can directly bind FIP200 Claw domain, but also can serve as an atypical ATG8-interacting motif to selectively recognize mammalian ATG8 family proteins (ATG8s). We determine the high-resolution crystal structures of ATG16L1 FIR in complex with FIP200 Claw and GABARAPL1, respectively, and elucidate the molecular mechanism underlying the interactions of ATG16L1 with FIP200 and ATG8s. To distinguish the precise contribution of FIP200 from ATG8s for binding to ATG16L1 FIR in autophagy, we develop a ATG16L1 mutant that can exclusively interact with ATG8s but not FIP200. Finally, using relevant cell-based functional assays, we demonstrate that the interaction of ATG16L1 with FIP200 is indispensable for the effective autophagic flux. In conclusion, our findings provide mechanistic insights into the interactions of ATG16L1 with FIP200 and ATG8s, and are valuable for further understanding the function of ATG16L1 in autophagy.
- State Key Laboratory of Chemical Biology, Shanghai Institute of Organic Chemistry, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: