Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Odagaki, Y., Murakami, Y., Takita, T., Mizutani, K., Mikami, B., Fujiwara, S., Yasukawa, K.(2024) Biochem Biophys Res Commun 733: 150728-150728
- PubMed: 39321488
- DOI: https://doi.org/10.1016/j.bbrc.2024.150728
- Primary Citation of Related Structures:
9JER, 9JF5, 9JFN - PubMed Abstract:
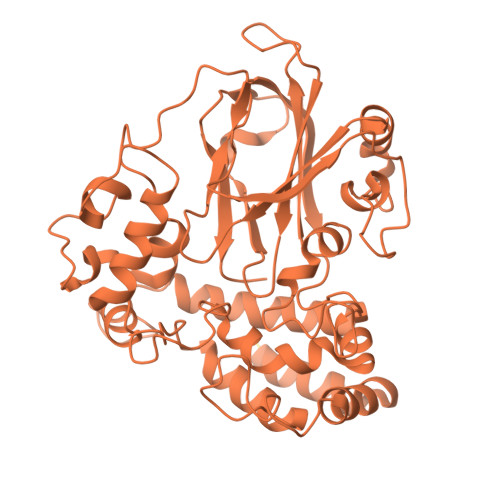
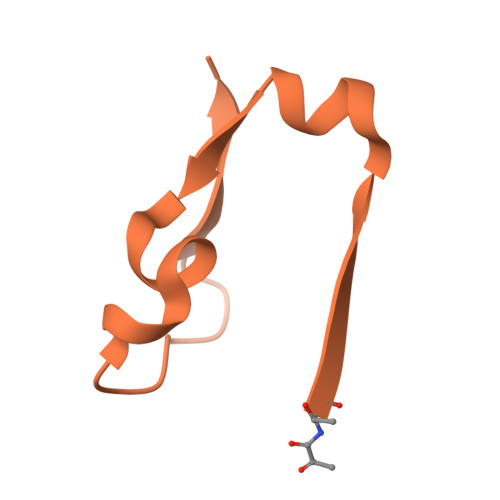
Agmatine, a natural polyamine also known as 4-aminobutyl-guanidine, is biosynthesized from arginine by decarboxylation. Aspergillus oryzae contains high amounts of agmatine, suggesting highly active arginine decarboxylase (ADC) in this organism. However, genome analysis revealed no ADC homolog in A. oryzae. A. oryzae strain RIB40 has six homologs of phosphatidylserine decarboxylase (PSD), an enzyme that synthesizes phosphatidyl ethanolamine from phosphatidylserine. We previously discovered that one of these homologs, AO090102000327, encodes arginine decarboxylase, which we named ADC1. In the present study, we determined the crystal structures of ligand-free, arginine-treated, and agmatine-treated ADC1 each at 1.9-2.15 Å resolution. Each structure contained four ADC1 molecules (chains A-D) in the asymmetric unit of the cell. Each ADC1 molecule is a heterodimer consisting of the N-terminal region (Asn60-Gly441) and C-terminal region (Ser442-Thr482). In the ligand-free ADC1, the N-terminus of Ser442 was modified to form a pyruvoyl group. In the arginine-treated ADC1, arginine was converted to agmatine, with the pyruvoyl group covalently bound to agmatine by forming a Schiff base. The same structure was observed in agmatine-treated ADC1. These results indicate that ADC1 is a pyruvoyl-dependent decarboxylase and unveils the reaction mechanism of ADC from A. oryzae.
- Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Sakyo-ku, Kyoto, 606-8502, Japan.
Organizational Affiliation: